Instructions for Side by Side Printing
- Print the notecards
- Fold each page in half along the solid vertical line
- Cut out the notecards by cutting along each horizontal dotted line
- Optional: Glue, tape or staple the ends of each notecard together
dna replication, damage, and repair irat 2
front 1 the replication/copying of cellular DNA occurs during the- | back 1 Synthesis phase (s phase) of the cell cycle |
front 2 The S phase is a necessary process to ensure that- | back 2 the instructions in DNA are faithfully passed on to the newly produced cells. |
front 3 in cell nuclei, DNA and protein complexes known as ____________ make up the ______________ | back 3 chromatin, chromosomes |
front 4 replication can ONLY occur on a- | back 4 single strand DNA template |
front 5 the main enzyme that catalyzes the formation of new DNA strands is- | back 5 DNA polymerase |
front 6 what is the function of DNA polymerase? | back 6 reads in 3' to 5' to form a complement strand in 5' to 3' |
front 7 DNA replication is ALWAYS | back 7 semi-conservative |
front 8 why is DNA replication semi- conservative? | back 8 uses the template of the original DNA + a newly synthesized strand. (each of the two daughter strands has half new DNA and half old DNA) |
front 9 the lagging strand is ? | back 9 the template strand |
front 10 the leading strand is? | back 10 the new strand |
front 11 a characteristic of eukaryotic DNA replication is that- | back 11 it is bidirectional, and starts in several locations at once |
front 12 DNA is copied at about- | back 12 50 base pairs per second |
front 13 What is the function of DNA helicase? | back 13 unwinds the two strands of DNA in a double helix |
front 14 what is the function of DNA primases? | back 14 synthesizes RNA primers to start DNA replication |
front 15 what is the function of DNA polymerases? | back 15 adds complementary bases to template strand, therefore creating a new DNA strand |
front 16 what is the function of exonucleases? | back 16 remove RNA fragments (Okazaki fragments), also act as a proofreader which removes nucleotides that are not a part of the double helix. (removes mismatched residues, edits DNA) |
front 17 What is the function of single-stranded DNA-binding proteins? | back 17 prevents DNA from binding back together |
front 18 What is the function of DNA ligase? | back 18 seals complementary strands |
front 19 what is the function of topoisomerases? | back 19 facilitates unwinding |
front 20 what is the function of telomerase? | back 20 synthesis of telomeres (caps the ends of a chromosome) |
front 21 What is the first 4 steps in DNA synthesis? | back 21 DNA helicase, DNA primase, DNA polymerase, Exonucleases |
front 22 What are the last four steps of DNA synthesis? | back 22 Single-stranded DNA binding proteins,DNA ligase, Topoisomerases,, Telomerases |
front 23 what base is found in RNA that is NOT found in DNA? | back 23 Uracil (U) |
front 24 RNA is a- | back 24 single strand |
front 25 Chain ELONGATION is carried out by- | back 25 DNA polymerases |
front 26 What is the specific function of DNA polymerases? | back 26 select the nucleotide that is added to the 3'-OH end of the growing chain and catalyze the formation of the phosphodiester bond. |
front 27 Exonucleases enhance the fidelity of DNA replication by- | back 27 rechecking the correctness of base pairing before proceeding with polymerization |
front 28 eukaryotic DNA replication is also- | back 28 a semi discontinuous process. |
front 29 why is DNA replication a semidiscontious process? | back 29 because the leading strand is synthesized CONTINUOUSLY, while the lagging strand is synthesized DISCONTINUOUSLY (Okazaki fragments) |
front 30 a NEW strand of DNA is always synthesized in the _________ direction | back 30 5' to 3' |
front 31 A strand is READ from the ___ end toward the __ end | back 31 3' , 5' |
front 32 ALL DNA polymerases function in the same manner in which they- | back 32 READ parental strand in 3' to 5' and SYNTHESIZE a complementary antiparallel new strand in 5' to 3' |
front 33 the leading strand is the one in which- | back 33 5' to 3' synthesis proceeds in the SAME direction as replication fork movement |
front 34 the lagging strand is the one in which- | back 34 DNA is 5' to 3' synthesized in short fragments in the opposite direction of the replication fork movement |
front 35 okazaki fragments are about ___ to ____ nucelotide long | back 35 100, 200 |
front 36 Overall chain growth occurs at the- | back 36 the base of the replication fork (but synthesis of the lagging strand occurs discontinuously in the opposite direction with exclusive 5'-3' polarity) |
front 37 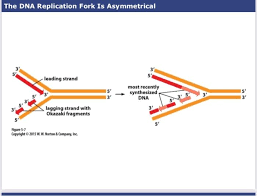 on the lagging strand, okazaki fragments are synthesized sequentially with fragments- | back 37 nearest to the replication fork being the most recently made |
front 38 what is annealing? | back 38 the ability of two complementary nucleic acids to align in an opposing orientation to form hydrogen bonds with bases of the complementary strand |
front 39 what keeps the strands of DNA replication protected until complementary strands are produced? | back 39 single-stranded DNA proteins |
front 40 why is DNA ligase so important? | back 40 it is important because it creates the final phosphodiester bond between adjacent nucleotides on a strand of DNA |
front 41 super twisting of DNA is removed by which enzyme? | back 41 topoisomerases |
front 42 what is the function of topoisomerase I ? | back 42 catalyzes breaks in only ONE strand of the double-stranded DNA |
front 43 what is the function of topoisomerase II? | back 43 catalyzes breaks in BOTH strands of the double stranded DNA |
front 44 both topoisomerases function similarly in that- | back 44 they allow the unwinding of the broken strand and catalyze the formation of new phosphodiester bonds |
front 45 Telomerase is an enzyme that- | back 45 helps maintain the telomere |
front 46 the telomere shortens as- | back 46 we get older |
front 47 what allows the cell to distinguish intact chromosomes from broken chromosomes and protect them from degradation | back 47 telomeres |
front 48 DNA damage can result from- | back 48 endogenous and exogenous causes |
front 49 most DNA damage is ________ before DNA is replicated | back 49 repaired |
front 50 Mutagenic agents are most effective during which phase of the cell cycle? | back 50 synthesis phase |
front 51 rate of endogenous mutations is termed- | back 51 the basal mutation rate |
front 52 what can contribute to errors in DNA replication? | back 52 spontaneous tautomeric shifts (changes from one natural form to another) |
front 53 what are examples of an exogenous agent? | back 53 ionizing radiation, hydrocarbons, oxidative free radicals, chemotherapy |
front 54 DNA repair is important because cells are- | back 54 continuously exposed to environmental mutagens and because cell mutations occur spontaneously in every cell during relplication |
front 55 cells use undamaged strands of DNA as a template to | back 55 correct the mistakes in DNA |
front 56 when both strands of DNA are damged, the cell uses- | back 56 the sister chromatid (2nd copy of DNA present in diploid cells) or an error-prone recovery system |
front 57 Define mismatch repair- | back 57 corrects the mismatches of normal bases that fail to maintain correct DNA pairing. (usually DNA polymerase is at fault) |
front 58 Proteins that can recognize a mismatch are | back 58 MSH2, MLH1, MSH6, PMS1, PMS2 genes |
front 59 mutations in MSH2, MLH1, MSH6, PMS1, PMS2 genes can lead to a predespotition in | back 59 hereditary nonpolyposis colon cancer (HNPCC) at a young age |
front 60 what corrects the spontaneous depurination and spontaneous deamination that happens to bases present in DNA | back 60 base excision repair |
front 61 spontaneous deamination of cytosine in DNA causes- | back 61 a conversion to uracil (which is only in RNA) |
front 62 Nucleotide excision repair is- | back 62 when an entire nucleotide is wrong |
front 63 nucleotide excision repair is specifically used to remove- | back 63 UV light-induced DNA damage, and damage from environmental chemicals |
front 64 UV light can form ________ from adjacent pyrimidine bases (C and G) in DNA | back 64 pyrimidine-pyrimidine dimers which can cause sunburn and skin cancer |
front 65 Nucleotide excision repair is also necessary to- | back 65 recognize chemically induced bulky additions to DNA that distort the shape of DNA which causes mutations |
front 66 what are two types of repair mechanisms that exist to correct DNA damage | back 66 homologous recombination and nonhomologous end joining |
front 67 homologous recombination- | back 67 takes advantage of sequence information available from the unaffected homologous chromosome for proper repair of breaks |
front 68 what proteins normally play a role in the homologous recombination process? mutations in these proteins/genes increase breast cancer risk. | back 68 BRCA 1 and 2 |
front 69 Nonhomologulos end joining is when- | back 69 the joining of ends even if the sequence is not similar. this is very error-prone and can cause mutation during repair |
front 70 oncogenes are- | back 70 mutated genes that can cause cancer and primarily encode cell cycle-related proteins |
front 71 what helps prevent uncontrolled dividing? | back 71 oncogenes and tumor suppressor genes help prevent uncontrolled dividing |