Campbell Biology Chapter 21 (powell_h)
1) For mapping studies of genomes, most of which were far along
before 2000, the three-stage method was often used. Which of the
following is the usual order in which the stages were performed,
assuming some overlap of the three?
A) genetic map, sequencing
of fragments, physical map
B) linkage map, physical map,
sequencing of fragments
C) sequencing of entire genome, physical
map, genetic map
D) cytogenetic linkage, sequencing, physical
map
E) physical map, linkage map, sequencing
Answer: B
2) What is the difference between a linkage map and a physical map?
A) For a linkage map, markers are spaced by recombination
frequency, whereas for a physical map they are spaced by numbers of
base pairs (bp).
B) For a physical map, the ATCG order and
sequence must be achieved; however, it does not for the linkage map.
C) For a linkage map, it is shown how each gene is linked to
every other gene.
D) For a physical map, the distances must be
calculable in units such as nanometers.
E) There is no
difference between the two except in the type of pictorial representation.
Answer: A
3) How is a physical map of the genome of an organism achieved?
A) using recombination frequency
B) using very
high-powered microscopy
C) using restriction enzyme cutting
sites
D) using sequencing of nucleotides
E) using DNA
fingerprinting via electrophoresis
Answer: C
4) Which of the following most correctly describes a shotgun
technique for sequencing a genome?
A) genetic mapping followed
immediately by sequencing
B) physical mapping followed
immediately by sequencing
C) cloning large genome fragments into
very large vectors such as YACs, followed by sequencing
D)
cloning several sizes of fragments into various size vectors, ordering
the clones, and then sequencing them
E) cloning the whole genome
directly, from one end to the other
Answer: D
5) The biggest problem with the shotgun technique is its tendency to
underestimate the size of the genome. Which of the following might
best account for this?
A) skipping some of the clones to be
sequenced
B) missing some of the overlapping regions of the
clones
C) counting some of the overlapping regions of the clones
twice
D) having some of the clones die during the experiment and
therefore not be represented
E) missing some duplicated sequences
Answer: E
6) What is metagenomics?
A) genomics as applied to a species
that most typifies the average phenotype of its genus
B) the
sequence of one or two representative genes from several species
C) the sequencing of only the most highly conserved genes in a
lineage
D) sequencing DNA from a group of species from the same
ecosystem
E) genomics as applied to an entire phylum
Answer: D
7) Which procedure is not required when the shotgun approach to
sequencing is modified as sequencing by synthesis, in which many small
fragments are sequenced simultaneously?
A) use of restriction
enzymes
B) sequencing each fragment
C) cloning each
fragment into a plasmid
D) ordering the sequences
E) PCR amplification
Answer: C
8) What is bioinformatics?
A) a technique using 3-D images of
genes in order to predict how and when they will be expressed
B)
a method that uses very large national and international databases to
access and work with sequence information
C) a software program
available from NIH to design genes
D) a series of search
programs that allow a student to identify who in the world is trying
to sequence a given species
E) a procedure that uses software to
order DNA sequences in a variety of comparable ways
Answer: B
9) What is proteomics?
A) the linkage of each gene to a
particular protein
B) the study of the full protein set encoded
by a genome
C) the totality of the functional possibilities of a
single protein
D) the study of how amino acids are ordered in a
protein
E) the study of how a single gene activates many proteins
Answer: B
10) Bioinformatics can be used to scan sequences for probable genes
looking for start and stop sites for transcription and for
translation, for probable splice sites, and for sequences known to be
found in other known genes. Such sequences containing these elements
are called
A) expressed sequence tags.
B) cDNA.
C)
multigene families.
D) proteomes.
E) short tandem repeats.
Answer: A
11) A microarray known as a GeneChip, with most now-known human
protein coding sequences, has been developed to aid in the study of
human cancer by first comparing two to three subsets of cancer
subtypes. What kind of information might be gleaned from this GeneChip
to aid in cancer prevention?
A) information about whether or not
a patient has this type of cancer prior to treatment
B) evidence
that might suggest how best to treat a person's cancer with
chemotherapy
C) data that could alert patients to what kind of
cancer they were likely to acquire
D) information about which
parent might have provided a patient with cancer-causing genes
E) information on cancer epidemiology in the United States or elsewhere
Answer: C
12) What is gene annotation in bioinformatics?
A) finding
transcriptional start and stop sites, RNA splice sites, and
ESTs
B) describing the functions of protein-coding genes
C)
describing the functions of noncoding regions of the genome
D)
matching the corresponding phenotypes of different species
E)
comparing the protein sequences within a single phylum
Answer: A
13) Why is it unwise to try to relate an organism's complexity with
its size or number of cells?
A) A very large organism may be
composed of very few cells or very few cell types.
B) A
single-celled organism, such as a bacterium or a protist, still has to
conduct all the complex life functions of a large multicellular
organism.
C) A single-celled organism that is also eukaryotic,
such as a yeast, still reproduces mitotically.
D) A simple
organism can have a much larger genome.
E) A complex organism
can have a very small and simple genome.
Answer: B
14) Fragments of DNA have been extracted from the remnants of extinct
woolly mammoths, amplified, and sequenced. These can now be used to
A) introduce into relatives, such as elephants, certain mammoth
traits.
B) clone live woolly mammoths.
C) study the
relationships among woolly mammoths and other wool-producers.
D)
understand the evolutionary relationships among members of related
taxa.
E) appreciate the reasons why mammoths went extinct.
Answer: D
15) If humans have 2,900 Mb, a specific member of the lily family has
120,000 Mb, and a yeast has ~13 Mb, why can't this data allow us to
order their evolutionary significance?
A) Size matters less than
gene density.
B) Size does not compare to gene density.
C)
Size does not vary with gene complexity.
D) Size is mostly due
to "junk" DNA.
E) Size is comparable only within phyla.
Answer: C
16) Which of the following is a representation of gene density?
A) Humans have 2,900 Mb per genome.
B) C. elegans has
~20,000 genes.
C) Humans have ~20,000 genes in 2,900 Mb.
D) Humans have 27,000 bp in introns.
E) Fritillaria has a
genome 40 times the size of a human.
Answer: C
17) Why might the cricket genome have 11 times as many base pairs as
that of Drosophila melanogaster?
A) The two insect species
evolved at very different geologic eras.
B) Crickets have higher
gene density.
C) Drosophila are more complex organisms.
D)
Crickets must have more noncoding DNA.
E) Crickets must make
many more proteins.
Answer: D
18) The comparison between the number of human genes and those of
other animal species has led to many conclusions, including
A)
the density of the human genome is far higher than in most other
animals.
B) the number of proteins expressed by the human genome
is far more than the number of its genes.
C) most human DNA
consists of genes for protein, tRNA, rRNA, and miRNA.
D) the
genomes of other organisms are most significantly smaller than the
human genome.
Answer: B
19) Barbara McClintock, who achieved fame for discovering that genes
could move within genomes, had her meticulous work ignored for nearly
four decades, but eventually won the Nobel Prize. Why was her work so
distrusted?
A) The work of women scientists was still not
allowed to be published.
B) Geneticists did not want to lose
their cherished notions of DNA stability.
C) There were too many
alternative explanations for transposition.
D) She allowed no
one else to duplicate her work.
E) She worked only with maize,
which was considered "merely" a plant.
Answer: B
20) What is the most probable explanation for the continued presence
of pseudogenes in a genome such as our own?
A) They are genes
that had a function at one time, but that have lost their function
because they have been translocated to a new location.
B) They
are genes that have accumulated mutations to such a degree that they
would code for different functional products if activated.
C)
They are duplicates or near duplicates of functional genes but cannot
function because they would provide inappropriate dosage of protein
products.
D) They are genes with significant inverted sequences.
E) They are genes that are not expressed, even though they have
nearly identical sequences to expressed genes.
Answer: E
21) What characteristic of short tandem repeat DNA makes it useful
for DNA fingerprinting?
A) The number of repeats varies widely
from person to person or animal to animal.
B) The sequence of
DNA that is repeated varies significantly from individual to
individual.
C) The sequence variation is acted upon differently
by natural selection in different environments.
D) Every racial
and ethnic group has inherited different short tandem repeats.
Answer: A
22) Alu elements account for about 10% of the human genome. What does
this mean?
A) Alu elements cannot be transcribed into RNA.
B) Alu elements evolved in very ancient times, before mammalian
radiation.
C) Alu elements represent the result of
transposition.
D) No Alu elements are found within individual
genes.
E) Alu elements are cDNA and therefore related to retrotransposons.
Answer: C
23) A multigene family is composed of
A) multiple genes whose
products must be coordinately expressed.
B) genes whose
sequences are very similar and that probably arose by duplication.
C) the many tandem repeats such as those found in centromeres
and telomeres.
D) a gene whose exons can be spliced in a number
of different ways.
E) a highly conserved gene found in a number
of different species.
Answer: B
24) Which of the following can be duplicated in a genome?
A)
DNA sequences above a minimum size only
B) DNA sequences below a
minimum size only
C) entire chromosomes only
D) entire
sets of chromosomes only
E) sequences, chromosomes, or sets of chromosomes
Answer: E
25) In comparing the genomes of humans and those of other higher
primates, it is seen that humans have a large metacentric pair we call
chromosome 2 among our 46 chromosomes, whereas the other primates of
this group have 48 chromosomes and any pair like the human chromosome
2 pair is not present; instead, the primate groups each have two pairs
of midsize acrocentric chromosomes. What is the most likely
explanation?
A) The ancestral organism had 48 chromosomes and at
some point a centric fusion event occurred and provided some selective
advantage.
B) The ancestral organism had 46 chromosomes, but
primates evolved when one of the pairs broke in half.
C) At some
point in evolution, human ancestors and primate ancestors were able to
mate and produce fertile offspring, making a new species.
D)
Chromosome breakage results in additional centromeres being made in
order for meiosis to proceed successfully.
E) Transposable
elements transferred significantly large segments of the chromosomes
to new locations.
Answer: A
26) Unequal crossing over during prophase I can result in one sister
chromosome with a deletion and another with a duplication. A mutated
form of hemoglobin, known as hemoglobin Lepore, is known in the human
population. Hemoglobin Lepore has a deleted set of amino acids. If it
was caused by unequal crossing over, what would be an expected
consequence?
A) If it is still maintained in the human
population, hemoglobin Lepore must be selected for in evolution.
B) There should also be persons born with, if not living long
lives with, an anti-Lepore mutation or duplication.
C) Each of
the genes in the hemoglobin gene family must show the same deletion.
D) The deleted gene must have undergone exon shuffling.
E)
The deleted region must be located in a different area of the
individual's genome.
Answer: B
27) When does exon shuffling occur?
A) during splicing of DNA
B) during mitotic recombination
C) as an alternative
splicing pattern in post-transcriptional processing
D) as an
alternative cleavage or modification post-translationally
E) as
the result of faulty DNA repair
Answer: C
28) What are genomic "hot spots"?
A) the locations
that correspond to most genetic diseases
B) the areas of a
genome that most often mutate due to environmental effects
C)
the locations that most often correspond with chromosomal breakpoints
D) the locations that correspond to most genetic diseases and
the locations that most often correspond with chromosomal breakpoints
E) the locations that correspond to most genetic diseases, the
areas of a genome that most often mutate due to environmental effects,
and the locations that most often correspond with chromosomal breakpoints
Answer: E
29) In order to determine the probable function of a particular
sequence of DNA in humans, what might be the most reasonable approach?
A) Prepare a knockout mouse without a copy of this sequence and
examine the mouse phenotype.
B) Genetically engineer a mouse
with a copy of this sequence and examine its phenotype.
C) Look
for a reasonably identical sequence in another species, prepare a
knockout of this sequence in that species, and look for the
consequences.
D) Prepare a genetically engineered bacterial
culture with the sequence inserted and assess which new protein is
synthesized.
E) Mate two individuals heterozygous for the normal
and mutated sequences.
Answer: C
30) Homeotic genes contain a homeobox sequence that is highly
conserved among very diverse species. The homeobox is the code for
that domain of a protein that binds to DNA in a regulatory
developmental process. Which of the following would you then expect?
A) that homeotic genes are selectively expressed over
developmental time
B) that a homeobox-containing gene has to be
a developmental regulator
C) that homeoboxes cannot be expressed
in nonhomeotic genes
D) that all organisms must have homeotic
genes
E) that all organisms must have homeobox-containing genes
Answer: A
31) Which of the following studies would not likely be characterized
as eco-devo?
A) the study of a particular species to see whether
or not it has developmental regulation
B) a study of the
assortment of homeotic genes in the zebra
C) a comparison of the
functions of a particular homeotic gene among four species of reptiles
D) studying the environmental pressures on developmental stages
such as the tadpole
E) a fossil comparison of organisms from the
Devonian era
Answer: C
32) A recent report has indicated several conclusions about
comparisons of our genome with that of Neanderthals. This report
concludes, in part, that, at some period in evolutionary history,
there was an admixture of the two genomes. This is evidenced by
A) some Neanderthal sequences not found in humans.
B) a
small number of modern H. sapiens with Neanderthal sequences.
C)
Neanderthal Y chromosomes preserved in the modern population of males.
D) mitochondrial sequences common to both groups.
Answer: B
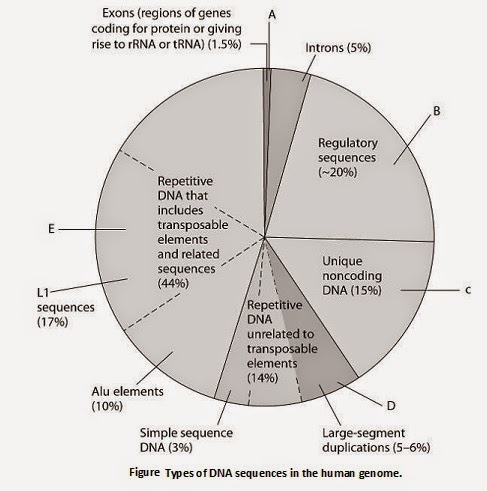
Figure 21.1 Types of DNA sequences in the human genome.
The pie chart in Figure 21.1 represents the relative frequencies
of the following in the human genome:
I. repetitive DNA
unrelated to transposons
II. repetitive DNA that includes
transposons
III. unique noncoding DNA
IV. introns and
regulatory sequences
V. exons
33) Which region is occupied by exons only (V)?
A) A
B) B
C) C
D) D
E) E
Answer: A

Figure 21.1 Types of DNA sequences in the human genome.
The pie chart in Figure 21.1 represents the relative frequencies
of the following in the human genome:
I. repetitive DNA
unrelated to transposons
II. repetitive DNA that includes
transposons
III. unique noncoding DNA
IV. introns and
regulatory sequences
V. exons
34) Which region includes Alu elements and LI sequences?
A) A
B) B
C) C
D) D
E) E
Answer: E

Figure 21.2 shows a diagram of blocks of genes on human chromosome 16
and the locations of blocks of similar genes on four chromosomes of
the mouse.
35) The movement of these blocks suggests that
A) during
evolutionary time, these sequences have separated and have returned to
their original positions.
B) DNA sequences within these blocks
have become increasingly divergent.
C) sequences represented
have duplicated at least three times.
D) chromosomal
translocations have moved blocks of sequences to other chromosomes.
E) higher mammals have more convergence of gene sequences
related in function.
Answer: D

Figure 21.2 shows a diagram of blocks of genes on human chromosome 16
and the locations of blocks of similar genes on four chromosomes of
the mouse.
36) Which of the following represents another example of the
same phenomenon as that shown in Figure 21.2?
A) the apparent
centric fusion between two chromosome pairs of primates such as chimps
to form the ancestor of human chromosome 2
B) the difference in
the numbers of chromosomes in five species of one genus of birds
C) the formation of several pseudogenes in the globin gene
family subsequent to human divergence from other primates
D) the
high frequency of polyploidy in many species of angiosperms
Answer: A
Multigene families include two or more nearly identical genes or
genes sharing nearly identical sequences. A classical example is the
set of genes for globin molecules, including genes on human
chromosomes 11 and 16.
37) How might identical and obviously duplicated gene sequences
have gotten from one chromosome to another?
A) by normal meiotic
recombination
B) by normal mitotic recombination between sister
chromatids
C) by transcription followed by recombination
D) by chromosomal translocation
E) by deletion followed by insertion
Answer: D
Multigene families include two or more nearly identical genes or
genes sharing nearly identical sequences. A classical example is the
set of genes for globin molecules, including genes on human
chromosomes 11 and 16.
38) Several of the different globin genes are expressed in
humans, but at different times in development. What mechanism could
allow for this?
A) exon shuffling
B) intron activation
C) pseudogene activation
D) differential translation of
mRNAs
E) differential gene regulation over time
Answer: E
39) Bioinformatics includes all of the following except
A)
using computer programs to align DNA sequences.
B) analyzing
protein interactions in a species.
C) using molecular biology to
combine DNA from two different sources in a test tube.
D)
developing computer-based tools for genome analysis.
E) using
mathematical tools to make sense of biological systems.
Answer: C
40) One of the characteristics of retrotransposons is that
A)
they code for an enzyme that synthesizes DNA using an RNA template.
B) they are found only in animal cells.
C) they generally
move by a cut-and-paste mechanism.
D) they contribute a
significant portion of the genetic variability seen within a
population of gametes.
E) their amplification is dependent on a retrovirus.
Answer: A
41) Homeotic genes
A) encode transcription factors that control
the expression of genes responsible for specific anatomical
structures.
B) are found only in Drosophila and other
arthropods.
C) are the only genes that contain the homeobox
domain.
D) encode proteins that form anatomical structures in
the fly.
E) are responsible for patterning during plant development.
Answer: A
42) Two eukaryotic proteins have one domain in common but are
otherwise very different. Which of the following processes is most
likely to have contributed to this similarity?
A) gene
duplication
B) RNA splicing
C) exon shuffling
D)
histone modification
E) random point mutations
Answer: C