front 1 The main reason scientists thought that proteins,
rather than DNA, were the carriers of genetic
material in the cell was:
-
their ability to be exported from the cell.
-
their presence within the nucleus.
-
their abundance within the cell.
-
their ability to self replicate within the cytoplasm.
-
the large number of possible amino acid
combinations.
| back 1 The large number of possible amino acid combinations. |
front 2 In the experiments of Griffith, the conversion of nonlethal
R-strain bacteria to lethal S-strain bacteria:
- supported the
case for proteins as the genetic material.
- was the result
of genetic mutation.
- was an example of conjugation.
- could not be reproduced by other researchers.
- was an
example of the genetic exchange known as transformation.
| back 2 was an example of the genetic
exchange known as transformation. |
front 3 The first experimenters to use Griffith's
transformation assay to identify the genetic material were:
- Hershey and
Chase.
- Meselson and Stahl.
- Watson and
Crick.
- Avery, MacLeod, and McCarty.
- Franklin and
Wilkins.
| back 3
Avery, MacLeod, and McCarty. |
front 4 Which of the following statements about DNA is
false?
- DNA is capable
of forming many different sequences.
- DNA contains the
sugar deoxyribose.
- DNA is double-stranded rather than
single-stranded.
- DNA is only found in eukaryotic cells.
- DNA contains thyme instead of uracil.
| back 4
DNA is only found in eukaryotic cells. |
front 5 The bacteriophages used in Alfred
Hershey's and Martha
Chase's experiments
showed that:
- DNA was injected
into bacteria.
- proteins were responsible for the
production of new viruses within the bacteria.
- proteins
were injected into bacteria.
- DNA remained on the outer
coat of bacteria.
- DNA and protein were injected into
bacteria.
| back 5
DNA was injected into bacteria. |
front 6 The two molecules that alternate to
form the backbone of a polynucleotide
chain are:
- Base and
phosphate.
- sugar and phosphate.
- cytosine and
guanine.
- base and sugar.
- adenine and
thymine.
| |
front 7
Chargaff determined that DNA from any source
contains about the same amount of guanine as __________.
- thymine
- cytosine
- adenine
- guanine
- uracil
| |
front 8 __________ used x-ray diffraction to provide images
of DNA.
- Crick and
Wilkins
- Franklin and Crick
- Watson and
Wilkins
- Franklin
- Watson and Crick
| |
front 9
X-ray diffraction studies are used to determine:
- the sequence of
amino acids in protein molecules.
- the type of chemical
under investigation.
- the distances between atoms of
molecules.
- the sequence of nucleic acids in nucleic acid
molecules.
- the wavelength of light emitted by
chemicals.
| back 9 the distances between atoms of molecules. |
front 10
X-ray crystallography showed that DNA:
- had the bases in
the center of the molecule.
- had the sugars and phosphates
on the outside of the molecule.
- was a helix.
- was
made of 2 strands.
- was a very long molecule.
| |
front 11 _________ determined the structure of the molecule DNA.
- Franklin and
Crick
- Crick and Wilkins
- Franklin
- Watson, Crick, and Wilkins
- Watson and Crick
| |
front 12
The information carried by DNA is incorporated in a code
specified by the:
- number of separate
strands of DNA.
- phosphodiester 0bonds of the DNA
strand.
- specific nucleotide sequence of the DNA
molecule.
- number of bases in a DNA strand.
- size of a
particular chromosome.
| back 12
specific nucleotide sequence of the DNA molecule. |
front 13
Why is DNA able to store large amounts of information?
- The nucleotides
can be altered to form many different letters in the sequence.
- It is capable of assuming a wide variety of shapes.
- Its nucleotides can be arranged in a large number of possible
sequences.
- It contains a large number of different
nucleotides.
- The sugar and phosphates can be arranged in many
different sequences.
| back 13 Its nucleotides can be arranged in a large number of
possible sequences. |
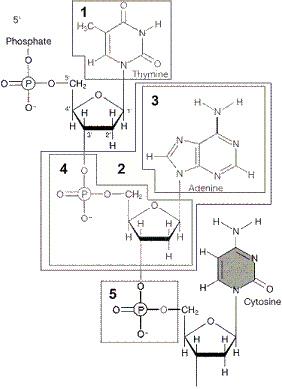
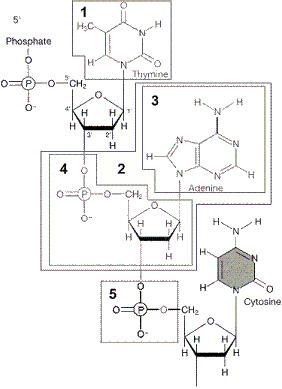
front 14
Figure 12-1 Use the figure to answer the
corresponding question(s). The portion of the molecule in box
5 of Figure 12-1 is:
- a hydrogen
bond.
- a nucleotide.
- a protein.
- a
pyrimidine.
- a phosphate.
| |
front 15
Figure 12-1 Use the figure to answer the
corresponding question(s). In Figure 12-1, the portion of the molecule
in box __________ is a pyrimidine.
- 1
- 1 and
3
- 3
- 4
- 3 and 4
| |
front 16
Figure 12-1 Use the figure to answer the
corresponding question(s). The portion of the molecule in
box 3 of Figure 12-1 is:
- a purine.
- a protein.
- a nucleotide.
- a pyrimidine.
- a sugar.
| |
front 17 Hydrogen bonds can form between guanine and
__________, and between adenine and __________.
- adenine;
guanine
- phosphate; sugar
- cytosine; thymine
- sugar; phosphate
- thymine; cytosine
| |
front 18 Two chains of DNA must run in __________
direction(s) and must be __________ if they are to
bond with each other.
-
parallel; complementary
-
parallel; uncomplementary
-
opposite; uncomplementary
-
the same; uncomplementary
-
antiparallel; complementary
| back 18
antiparallel; complementary |
front 19 Which of the following nucleotide sequences represents the
complement to the DNA strand 5¢ - AGATCCG- 3¢?
- 3¢ - AGATCCG-
5¢
- 5¢ - CTCGAAT- 3¢
- 5¢ - AGATCCG- 3¢
- 3¢
- TCTAGGC- 5¢
- 3¢ - CTCGAAT- 5¢
| |
front 20
When DNA copies itself,
- the two strands
stay intact as a double helix and the double helix serves as a
template for a new double helix.
- the two strands separate
and one of the two serves as a template.
- the two strands
separate and both strands serve as templates.
- the DNA
molecule completely hydrolyzes down to its individual nucleotides
which then reorganize to form two new DNA molecules.
- nucleotides rearrange themselves adn pair together in all
possible combinations.
| back 20 the two strands separate and both strands
serve as templates. |
front 21 Which of the following best describes semiconservative replication?
-
The translation of a DNA molecule into a complementary strand
of RNA.
-
A DNA molecule consists of one parental strand and one new
strand.
-
The replication of DNA takes place at a defined period in the
cell cycle.
-
The replication of DNA never takes place with 100% accuracy.
-
The number of DNA molecules is doubled with every other
replication.
| back 21 A DNA molecule consists of one parental
strand and one new strand. |
front 22 The final product of DNA replication is:
- the enzymes needed
for further processes, such as DNA polymerase.
- a wide
variety of proteins.
- two DNA molecules, each of which
contains one new and one old DNA strand.
- DNA
fragments.
- mRNA, tRNA, and rRNA molecules.
| back 22
two DNA molecules, each of which
contains one new and one old DNA strand. |
front 23 Who first confirmed that the replication of DNA was semiconservative?
- Watson and
Crick
- Watson, Crick, and Wilkin
- Chargaff and
Hershey
- Avery and Griffith
- Meselson and
Stahl
| |
front 24 If DNA replication rejoined the 2 parental strands,
it would be termed:
- parental.
- semiconservative.
- conservative.
- gradient.
- dispersive.
| |
front 25
Meselson and Stahl separated DNA from different
generations using:
-
gel electrophoresis.
-
density gradient centrifugation.
-
None of these.
-
an electron microscope.
-
differential radioisotope labeling.
| back 25
density gradient centrifugation |
front 26 When a DNA molecule containing a wrong base at one
location in one strand is replicated:
- the mutation is
copied into one of the two daughter molecules.
- the
mutation is ignored by the DNA polymerase enzyme.
- the
mutation is corrected by the DNA polymerase enzyme.
- the
mutation is copied into both of the daughter molecules.
- the replication is stopped.
| back 26 the mutation is copied into one of
the two daughter molecules. |
front 27 Which of the following causes the unwinding of the DNA double helix?
- RNA polymerase
- DNA polymerase
- RNA primer
- DNA helicase
- primosome
| |
front 28
A replication fork:
- is a site where one
DNA strand serves as a template, but the other strand is not
replicated.
- is only seen in bacterial cells.
- is
created by the action of the enzyme RNA polymerase.
- is only
seen in prokaryotic chromosomes.
- is a Y-shaped structure
where both DNA strands are replicated simultaneously.
| back 28 is a Y-shaped structure where both
DNA strands are replicated simultaneously. |
front 29 In replication, once the DNA strands have
been separated, reformation of the double
helix is prevented by:
- DNA polymerases.
- GTP.
- DNA helicase enzyme.
- single-strand binding
proteins.
- ATP.
| back 29
single-strand binding proteins. |
front 30
Enzymes called __________ form breaks in the
DNA molecules to prevent the formation of
knots in the DNA helix during replication.
- RNA polymerases
- single-strand binding proteins
- DNA ligases
- DNA
polymerases
- topoisomerases
| |
front 31 What prevents knot formation in replicating DNA?
-
scaffolding proteins
-
histones
-
topoisomerases
-
protosomes
-
chromatin
| |
front 32 Which of the following adds new nucleotides to a
growing DNA chain?
- RNA polymerase
- primase
- DNA helicase
- RNA primer
- DNA
polymerase
| |
front 33
Why does DNA synthesis only proceed in the 5¢ to 3¢ direction?
- Because that is the
direction in which the two strands of DNA unzip.
- Because
the 3¢ end of the polynucleotide molecule is more electronegative
than the 5¢ end.
- Because that is the only direction that
the polymerase can be oriented.
- Because DNA polymerases can
only add nucleotides to the 3¢ end of a polynucleotide strand.
- Because the chromosomes are always aligned in the 5¢ to 3¢
direction in the nucleus.
| back 33 Because DNA polymerases can only add nucleotides to the 3¢
end of a polynucleotide strand. |
front 34 The 5¢ end of each Okazaki fragment begins with:
-
a DNA primer binding to the template DNA.
-
the same RNA primer that began synthesis on the leading
strand.
-
a separate RNA primer.
-
DNA polymerase binding to the template DNA.
-
a small DNA primer.
| |
front 35 DNA primase is the enzyme responsible for:
- unwinding the DNA
double strand to allow DNA polymerase access to the template
DNA.
- introducing nicks into the DNA double strand in
order to prevent the formation of knots.
- hydrolyzing ATP
to facilitate DNA unwinding.
- forming a replication fork in
the DNA double helix.
- making short strands of RNA at the
site of replication initiation.
| back 35
making short strands of RNA at the site of
replication initiation. |
front 36 The DNA strand that is replicated smoothly and
continuously is called the:
- alpha strand.
- first strand.
- primary strand.
- lagging
strand.
- leading strand.
| |
front 37 In DNA replication, the lagging strand:
- is not synthesized
until the synthesis of the leading strand is completed.
- pairs with the leading strand by complementary base
pairing.
- is synthesized as a complementary copy of the
leading strand.
- is synthesized as a series of Okazaki
fragments.
- is made up entirely of RNA primers.
| back 37 is synthesized as a series of Okazaki fragments. |
front 38
Okazaki fragments are joined together by:
- DNA polymerase.
- RNA polymerase.
- RNA ligase.
- DNA ligase.
- primase.
| |
front 39
How is the chromosome of a bacterial cell replicated?
- The linear DNA
molecule is replicated from one origin of replication
bidirectionally.
- The circular DNA molecule is replicated
from one origin of replication bidirectionally.
- The
circular DNA molecule is replicated from one origin of replication
unidirectionally.
- The linear DNA molecule is replicated
from multiple origins of replication bidirectionally.
- The
circular DNA molecule is replicated from multiple origins of
replication bidirectionally.
| back 39 The circular DNA molecule is replicated from
one origin of replication bidirectionally. |
front 40
How are the chromosomes of a eukaryote cell replicated?
- The linear DNA
molecules are replicated from multiple origins of replication
bidirectionally.
- The circular DNA molecules are replicated
from multiple origins of replication bidirectionally.
- The
circular DNA molecules are replicated from one origin of replication
bidirectionally.
- The linear DNA molecules are replicated from
one origin of replication bidirectionally.
- The linear DNA
molecules are replicated from one origin of replication
unidirectionally.
| back 40 The linear DNA molecules are replicated from
multiple origins of replication bidirectionally. |
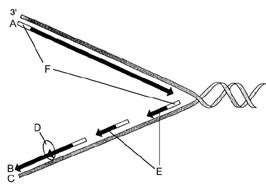
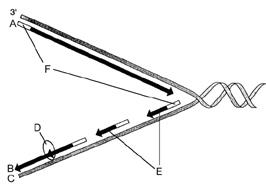
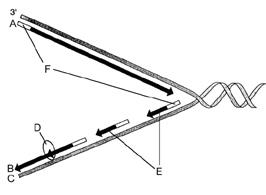
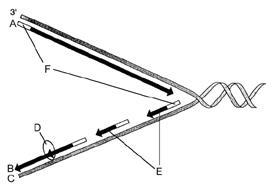
front 41
Figure 12-2 Use the figure to answer the
corresponding question(s). Refer to Figure 12-2. The correct
designation for the DNA strand labeled C is:
- the leading
strand.
- 3¢.
- Okazaki fragments.
- a strand
serving as a template.
- polymerase.
| back 41 a strand serving as a template. |
front 42
Figure 12-2 Use the figure to answer the
corresponding question(s). Refer to Figure 12-2. The segments
labeled F are responsible for:
- unwinding the DNA double
helix.
- synthesizing the leading strand.
- initiating
DNA synthesis.
- linking short DNA segments.
- forming
the replication fork.
| back 42
initiating DNA synthesis. |
front 43
Figure 12-2 Use the figure to answer the
corresponding question(s). Refer to Figure 12-2. The enzyme
represented by the letter D is responsible for:
- unwinding the DNA double
helix.
- forming the replication fork.
- linking short
DNA segments.
- forming nucleosomes.
- synthesizing
the leading strand.
| back 43
linking short DNA segments. |
front 44
Figure 12-2 Use the figure to answer the
corresponding question(s). Refer to Figure 12-2. The
structures represented by the letter E are called:
- replication forks.
- nucleosomes.
- DNA polymerases.
- leading
fragments
- Okazaki fragments.
| |
front 45 Which of the following statements concerning nucleotide
excision repair is FALSE?
- It is implicated in
xeroderma pigmentosum.
- It involves a DNA polymerase.
- It involves a nuclease.
- It involves DNA ligase.
- It is a type of mismatch repair.
| back 45
It is a type of mismatch repair. |
front 46 __________, the ends of eukaryotic chromosomes, shorten with
every cell replication event.
- Primosomes
- Centromeres
- Nucleosomes
- Kinetochores
- Telomeres
| |
front 47 The ends of eukaryotic chromosomes can be lengthened by:
-
primase.
-
apoptosis.
-
DNA polymerase.
-
reverse transcriptase.
- telomerase.
| |
front 48 When cultured normal human cells were infected with a
virus that carried the
genes that coded for a subunit of telomerase:
- the cells underwent more
cell divisions than normal.
- the cells underwent gene
expression more vigorously.
- the cells died almost
immediately.
- the cells underwent fewer cell divisions than
normal.
- the cell cycle shortened.
| back 48
the cells underwent more cell divisions than normal. |
front 49
Cancerous cells differ from noncancerous cells in
that cancerous cells:
- divide only 20 to 30
times.
- can maintain telomere length as
they divide.
- have lost the ability to resist
apoptosis.
- have reduced levels of telomerase.
- lack
telomeres
| back 49
can maintain telomere length as they divide. |
front 50
Mismatch repair occurs when
- enzymes repair DNA
lesions (deformed DNA) caused by ultraviolet radiation
- the
two strands of DNA in a double helix are parallel instead of
antiparallel
- an art student who has been placed in General
Biology I for science majors is moved to a nonscience majors biology
class.
- enzymes remove incorrectly-paired nucleotides.
- chromosome pairs are separated during meiosis
| back 50
enzymes remove incorrectly-paired nucleotides. |