Instructions for Side by Side Printing
- Print the notecards
- Fold each page in half along the solid vertical line
- Cut out the notecards by cutting along each horizontal dotted line
- Optional: Glue, tape or staple the ends of each notecard together
Ch.11
front 1 Enzymes are catalyst and | back 1 speed up reactions and are constantly moving |
front 2 Active site conformation changes can: | back 2 1. Assist substrate binding 2. Bring catalytic groups into position 3. Assist in bond making and bond breaking 4. Facilitate conversion of substrate to product |
front 3 Enzymes differ from ordinary chemical catalysts in | back 3 reaction rate, reaction conditions, reaction specificity, and regulation |
front 4 The unique physical and chemical properties of the_____limit an enzymes activity to specific substrates and reactions | back 4 active sites |
front 5 Some enzymes require | back 5 metal ions or organic cofactors |
front 6 Enzymes catalyze________reactions, causing them to proceed at extraordinarily rapid rates | back 6 thermodynamically favored |
front 7 what word is used to describe rates | back 7 kinetic |
front 8 The catalyzed and uncatalyzed rate: | back 8 catalyzed: 3.0x104sec^-1 uncatayzed: 3.0x10-10sec^-1 |
front 9 How to calculate the ratio for catalytic power? | back 9 3.0x104/3.0x10-10=1.0x1014 |
front 10 Any chemical reaction in which the oxidation numbers of the atoms are changed | back 10 oxidation-reduction(redox) reaction |
front 11 What is the classification of an oxidation-reduction reaction? | back 11 oxidoreductases |
front 12 What is the classification of a transfer of functional groups? | back 12 transferases |
front 13 What is the classification of a hydrolysis reactions? | back 13 hydrolases |
front 14 What is the classification of a group elimination to form double bonds? | back 14 lyases |
front 15 What is the classification of isomerization? | back 15 isomerases |
front 16 What is the classification of a bond formation coupled with ATP hydrolysis? | back 16 Ligases |
front 17 The_______from one molecule (the donor) to another (the acceptor) | back 17 transfer of functional groups |
front 18 A chemical compound by a reaction with water. Breaks down a variety of polymers, including proteins, carbs, fats, and nucleic acids | back 18 Hydrolysis |
front 19 Catalyze the breaking of various chemical bonds by means other than hydrolysis and oxidation, often forming new double bonds | back 19 Lyases |
front 20 Structural rearrangement of isomers (same molecular weight, but diff. structural formulas) | back 20 Isomerization |
front 21 Reaction joining of two large molecules by forming a new chemical bond | back 21 Ligases |
front 22 the selectivity of enzymes for their substrates | back 22 Enzyme specificity |
front 23 Exquisite stereospecificity is observed | back 23 for some enzymes |
front 24 what are cofactors? | back 24 are non-protein groups that are needed in active sites to carry out functions that amino acid side chains cannot |
front 25 Functional groups of proteins facilitate | back 25 -acid-base reactions - transient(short-lived)covalent bonds -Charge-charge interactions |
front 26 Cofactors have | back 26 metal ions and coenzymes |
front 27 Coenzymes have | back 27 Cosubstrates and prosthetic groups |
front 28 Transiently associates with enzymes so that it functions as a substrate | back 28 cosubstrates |
front 29 permanently(often covalently) attached to enzymes | back 29 Prosthetic groups |
front 30 ________are catalytically active enzymes with its cofactor complex | back 30 Holoenzymes |
front 31 ______enzyme without the cofactor | back 31 Apoenzyme |
front 32 _______must be regenerated for completion of a "catalytic cycle" | back 32 Coenzymes |
front 33 What are the five principles of regulated enzyme activity? | back 33 1. Allosteric control 2. Multiple forms of enzymes 3. Reversible covalent modification 4. Proteolytic activation 5. Controlling the amount of enzyme present |
front 34 Are inactive precursors of enzymes | back 34 zymogens |
front 35 Proteolytic cleavage produces the active enzyme | back 35 chymotrypsinogen---->chymotrypsin |
front 36 are non-protein components essential to enzyme activity | back 36 Enzyme cofactors and coenzymes |
front 37 An enzyme provides a lower-energy pathway from substrate to product but | back 37 does not affect the overall free energy change for the reaction |
front 38 The active sites of enzymes bind the transition state of the reaction more tightly than they____ | back 38 bind to the substrate |
front 39 The transition state sits at the _____ of the energy profile in the energy diagram | back 39 apex |
front 40 the reaction rate is proportional to the | back 40 concentration of reactant molecules that reached the transition-state energy |
front 41 The higher the delta G^+-, | back 41 the slower the reaction |
front 42 Decreasing delta G^+-, | back 42 increase the reaction rate (speeds up the reaction) |
front 43 The catalytic role of an enzyme is to reduce the energy barrier between | back 43 substrate s and transition state X^+- |
front 44 Rate acceleration by an enzyme means that the energy barrier between | back 44 ES and EX^+- must be smaller than the barrier between S & X^+- |
front 45 The enzyme must stabilize the EX^+- transition state | back 45 more than it stabilizes ES |
front 46 Binding cannot be too tight because the goal is to make | back 46 the energy barrier between ES EX^+- small |
front 47 RAising the starting energy of ES to a more positive delta G, | back 47 will increase the catalyzed rate |
front 48 The ES complex is a more highly ordered | back 48 for low-entropy state for the substrate |
front 49 Amino-acid side chains that can donate or accept protons can participate in chemical reactions as acid and base catalysts | back 49 Acid/base catalysis |
front 50 Groups can catalyze reactions through the transient formation of covalent bonds with the substrate | back 50 Nucleophilic attack |
front 51 The unique electronic properties of a metal ion facilitate the reaction | back 51 Metal ion catalysis |
front 52 Enzymes accelerate reaction by bringing reacting group together and orienting them for reaction | back 52 Proximity and Orientation |
front 53 Significantly lowers the activation energy for a reaction | back 53 Transition state stabilization |
front 54 A binding pocket determines the | back 54 substrate specificity of the various serine proteases |
front 55 the catalytically active__________ residues of serine proteases were identified by chemical labeling and structural analysis | back 55 Ser, His, and Asp |
front 56 what is step 1 of the mechanism of serine protease? | back 56 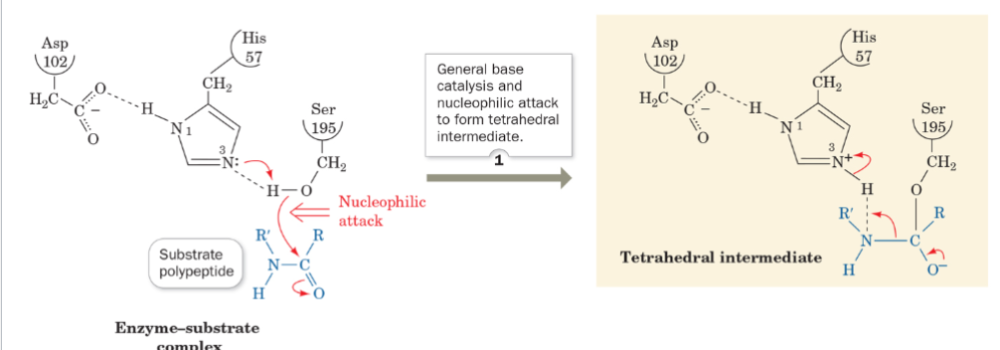 General base catalysis, nucleophilic attack to form tetrahedral intermediate |
front 57 what is step 2 of the mechanism of serine protease? | back 57 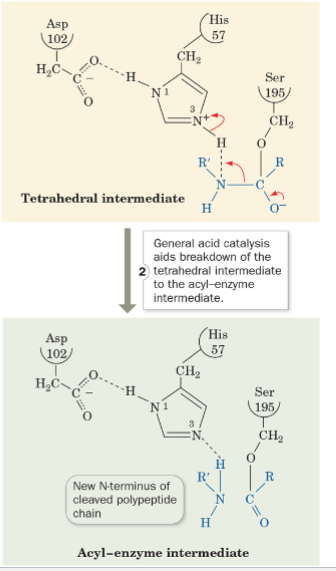 General Acid Catalysis aids breakdown of |
front 58 what is step 3 of the mechanism of serine protease? | back 58 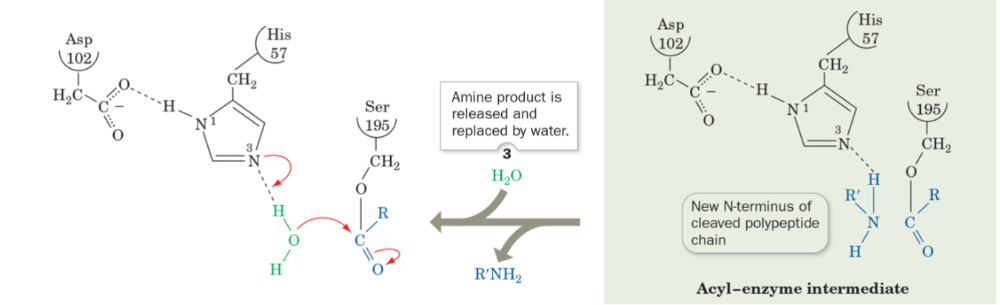 Amine product is released and replaced by water |
front 59 what is step 4 of the mechanism of serine protease? | back 59 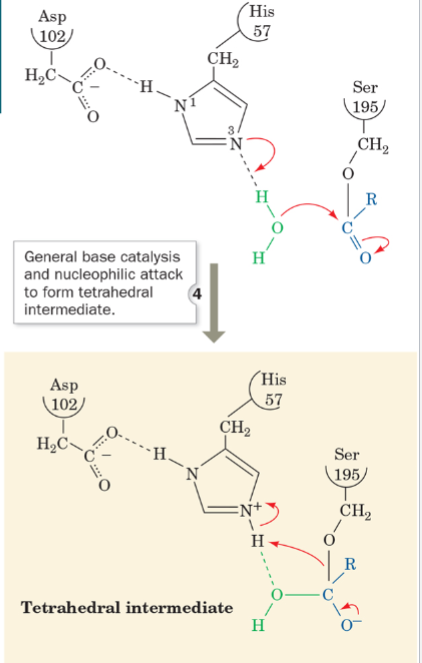 General Base Catalysis, Nucleophilic |
front 60 what is step 5 of the mechanism of serine protease? | back 60 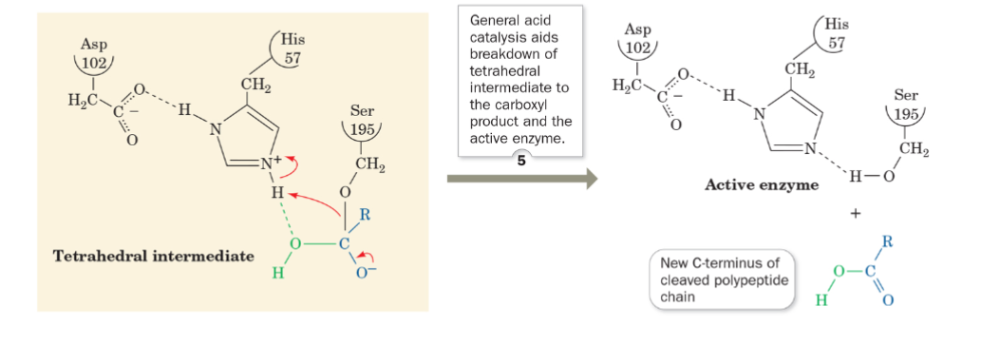 General acid catalysis aids breakdown of tetrahedral intermediate to the carboxyl product and the active enzyme |
front 61 Raising the energy of ES is accomplished by | back 61 A) loss of entropy B) destabilization of ES complex by -desolvation -strain/distortion |
front 62 Entropy loss is caused by the | back 62 formation of the ES complex. This complex is highly ordered, low-entropy state for a substrate |
front 63 Desolvation is the result of | back 63 water loss which raises the energy of the ES complex |
front 64 Destabilization is | back 64 provoking repulsion based on charges |
front 65 Proximity and Orientation | back 65 Asp 102 functions only to orient His57 |
front 66 Acid/base catalysis | back 66 His57 acts as general acid and base |
front 67 Nucleophilic attack | back 67 Ser195 formas a transient covalent bond with peptide to be cleaved trigonal to tetrahedral |
front 68 transition state-stabilization | back 68 tetrahedral oxyanion intermediate is stabilized by the backbone N-H of Gly193 and Ser195 the Oxyanion Hole |
front 69 Segments of RNA that display enzyme activity | back 69 riboenzymes |