Instructions for Side by Side Printing
- Print the notecards
- Fold each page in half along the solid vertical line
- Cut out the notecards by cutting along each horizontal dotted line
- Optional: Glue, tape or staple the ends of each notecard together
exam 4 BIOL213
front 1 Consider the process that a cell uses to replicate its double-stranded DNA before undergoing cell division. Which statement describes the DNA in the resulting daughter cells? | back 1 The double helix in each daughter cell consists of one parent strand and one newly synthesized strand. |
front 2 When is homologous recombination, which can flawlessly repair double-strand DNA breaks, most likely to occur? | back 2 after the cell’s DNA has been replicated |
front 3 DNA mismatch repair typically corrects what percentage of replication errors? | back 3 99% |
front 4 What is true of eukaryotic mRNAs? | back 4 They are translated after they are exported from the nucleus. |
front 5 An RNA chain is synthesized in which direction? | back 5 5′-to-3′ only |
front 6 In eukaryotes, which parts of a gene are transcribed into RNA? | back 6 introns and exons |
front 7 How many different aminoacyl-tRNA synthetases do most organisms have? | back 7 one for each amino acid |
front 8 What is false regarding codons in mRNA molecules? | back 8 Some codons code for more than one amino acid. |
front 9 It is thought that, early in the history of life, RNAs not only stored information but acted as catalysts in cells. Which reaction is catalyzed by a ribozyme in present-day cells? | back 9 peptide bond formation during translation |
front 10 Which of the following statements is NOT true about liver cells and kidney cells in the same organism? | back 10 They contain different genes. |
front 11 Which of the following would be the best method for determining which genes are being transcribed in a particular cell type? | back 11 RNA sequencing |
front 12 In eukaryotes, where do transcription regulators bind? | back 12 upstream, downstream, or within the genes they control |
front 13 What is an operon? | back 13 a set of genes transcribed as a single mRNA from a single promoter |
front 14 Which is NOT an example of epigenetic inheritance? | back 14 the inheritance of a single point mutation in a gene |
front 15 Which does NOT affect gene expression at the post-transcriptional level? | back 15 DNA methylation |
front 16 A scientist prepared 1.2 liters of a 250 mM sodium chloride (NaCl)
solution. However, after preparing the solution (but not using it),
she determines that the experiment setup needs 20% more solution than
originally planned. Because it is critical that the solution
concentration be exactly 250 mM NaCl, she decides to:1. Increase the
volume of the solution to 1.5 liters. | back 16 She needs to add 4.38 g NaCl to the existing solution and then complete the volume to 1.5 liters. |
front 17 A chemist needs to prepare 350 ml of a 100 mM potassium nitrate
(KNO3) solution. However, after preparing the solution, the chemist
realizes that the experiment requires the final concentration to be
150 mM. Because it is critical that the solution concentration be
exactly 150 mM KNO3, the chemist decides to: Considering the molecular weight of KNO3 is 101.10 g/mol, select the correct answer below: | back 17 The chemist needs to add 4.08 g KNO3 to the existing solution and then complete the volume to 0.5 liters. |
front 18 A scientist initially prepared 2.0 liters of a 150 mM potassium
chloride (KCl) solution. After reviewing the experiment setup, she
realizes that she now needs to increase the solution volume by 30%,
maintaining the same concentration of 150 mM. To ensure
accuracy: | back 18 She needs to complete the volume to 2.6 liters and then add 6.72 g KCl to the solution. |
front 19 A chemist prepared 750 ml of a 400 mM magnesium sulfate (MgSO4)
solution. Upon realizing that the experiment now requires 20% more
solution, the chemist decides to increase the volume while keeping the
concentration exactly at 400 mM. The chemist: | back 19 8.64 g |
front 20 A biologist is preparing a 500 ml solution of 0.5 M calcium chloride
(CaCl2) for an experiment. Later, she realizes that she needs an
additional 40% volume, while the concentration must remain exactly 0.5
M. The solution’s volume is increased to 700 ml. To correct the
concentration: | back 20 11.10 g |
front 21 A technician prepared 1 liter of a 300 mM lithium chloride (LiCl)
solution. The experiment calls for an additional 50% volume increase,
while the concentration must remain exactly 300 mM. The technician
decides to increase the volume to 1.5 liters. To correct the
concentration: | back 21 6.36 g |
front 22 What is the function of primase in RNA primer synthesis? | back 22 It uses ribonucleoside triphosphates to create RNA primers. |
front 23 How does primase differ from DNA polymerase? | back 23 Primase does not require a base-paired 3’ end to start synthesis. |
front 24 What type of chemical modification commonly results from UV radiation
and can | back 24 Formation of thymine dimers |
front 25 Which of the following mutations is caused by the deamination of cytosine? | back 25 Cytosine is converted into uracil |
front 26 If a guanine base undergoes oxidation, what type of mutation can occur if the damage is not repaired? | back 26 conversion of guanine into 8-oxoguanine |
front 27 What happens when 8-oxoguanine is mispaired during DNA replication? | back 27 It pairs with adenine, leading to a G:C to T:A transversion |
front 28 Which of the following is a consequence of the methylation of cytosine in DNA? | back 28 Increased likelihood of spontaneous deamination to thymine |
front 29 How does deamination of adenine lead to mutations if left unrepaired? | back 29 Adenine is converted to hypoxanthine, leading to mispairing with cytosine |
front 30 Which of the following best describes the mutation caused by the
oxidation of | back 30 It causes thymine to form 5-hydroxymethyluracil, which can pair
incorrectly |
front 31 Which of the following is true regarding the formation of abasic
sites (loss of a | back 31 DNA polymerase inserts random bases opposite the abasic site, leading to mutations |
front 32 What is the primary function of the excision step in DNA repair? | back 32 Removal of the damaged DNA strand |
front 33 Which enzyme is primarily responsible for the re-synthesis step in DNA repair? | back 33 DNA polymerase |
front 34 What role does DNA ligase play in the DNA repair process? | back 34 It seals the nick between the newly synthesized DNA and the existing strand. |
front 35 Which of the following best describes nucleotide excision repair (NER)? | back 35 It repairs damage caused by thymine dimers and other bulky lesions. |
front 36 What is the primary function of base excision repair (BER)? | back 36 It removes and replaces damaged bases like oxidized or deaminated bases. |
front 37 Which enzyme recognizes and removes the damaged base in base excision repair? | back 37 DNA glycosylase |
front 38 During mismatch repair (MMR), what recognizes the mismatch and distinguishes the newly synthesized strand from the template strand? | back 38 MutS and MutL proteins |
front 39 How does the repair of double-strand breaks by homologous recombination (HR) occur? | back 39 It uses the sister chromatid as a template to accurately repair the break. |
front 40 What is the function of DNA helicase during DNA replication? | back 40 It unwinds the DNA double helix. |
front 41 Which enzyme synthesizes the RNA primer required for DNA replication? | back 41 Primase |
front 42 In what direction does DNA polymerase synthesize new DNA strands? | back 42 5’ to 3’ direction |
front 43 What is the name of the fragments synthesized on the lagging strand
during DNA | back 43 Okazaki fragments |
front 44 Which enzyme is responsible for joining Okazaki fragments together? | back 44 DNA ligase |
front 45 What is the role of single-stranded binding proteins (SSBs) in DNA replication? | back 45 They stabilize the single-stranded DNA and prevent reannealing. |
front 46 Which enzyme relieves the tension ahead of the replication fork by breaking and rejoining the DNA strands? | back 46 Topoisomerase |
front 47 What is the leading strand in DNA replication? | back 47 The strand synthesized in the 5’ to 3’ direction toward the replication fork |
front 48 Which enzyme is primarily responsible for adding new nucleotides to the growing DNA strand during replication? | back 48 DNA polymerase |
front 49 How does replication of the lagging strand differ from replication of the leading strand? | back 49 The lagging strand is synthesized in short fragments that are later joined together, while the leading strand is synthesized continuously. |
front 50 What is the role of the origin of replication in DNA replication? | back 50 It is the specific sequence of DNA where replication begins. |
front 51 What is the main function of telomerase? | back 51 It adds repetitive nucleotide sequences to the ends of chromosomes. |
front 52 Why is telomerase important for the replication of eukaryotic chromosomes? | back 52 It extends the 3’ ends of chromosomes, preventing their shortening during |
front 53 Which type of cells typically express high levels of telomerase? | back 53 Cancer cells |
front 54 What happens to chromosomes in most somatic cells when telomerase is not | back 54 The telomeres shorten with each round of cell division. |
front 55 Which of the following is a component of the telomerase enzyme? | back 55 An RNA template that is complementary to the telomere sequence |
front 56 What is the correct sequence of information flow according to the
central dogma | back 56 DNA → RNA → Protein |
front 57 What process describes the conversion of DNA into RNA? | back 57 Transcription |
front 58 Which enzyme is responsible for synthesizing RNA from a DNA template? | back 58 RNA polymerase |
front 59 What is the process by which RNA is translated into a protein? | back 59 Translation |
front 60 Which type of RNA carries the genetic information from DNA to the
ribosome for | back 60 Messenger RNA (mRNA) |
front 61 Which cellular organelle is the site of protein synthesis? | back 61 Ribosome |
front 62 What is the role of transfer RNA (tRNA) during translation? | back 62 It brings amino acids to the ribosome based on the mRNA sequence. |
front 63 Which of the following best describes the genetic code? | back 63 It is universal and redundant, with multiple codons encoding the same amino |
front 64 Which codon serves as the start signal for translation in most organisms? | back 64 AUG |
front 65 What is the process by which RNA is converted back into DNA in some viruses? | back 65 Reverse transcription |
front 66 What is the primary function of messenger RNAs (mRNAs)? | back 66 They code for proteins. |
front 67 Which type of RNA forms the core of the ribosome’s structure and
catalyzes | back 67 Ribosomal RNAs (rRNAs) |
front 68 Which RNA type is primarily involved in regulating gene expression? | back 68 MicroRNAs (miRNAs) |
front 69 What role do transfer RNAs (tRNAs) play in protein synthesis? | back 69 They serve as adaptors between mRNA and amino acids. |
front 70 Which type of RNA provides protection from viruses and
proliferating | back 70 Small interfering RNAs (siRNAs) |
front 71 What is one function of long noncoding RNAs (lncRNAs)? | back 71 They act as scaffolds and serve other diverse functions. |
front 72 Which of the following RNA types is involved in RNA splicing, gene
regulation, | back 72 Other noncoding RNAs |
front 73 Which RNA type does not directly code for proteins but instead serves
a | back 73 Ribosomal RNAs (rRNAs)v |
front 74 Which type of RNA can regulate gene expression by interfering with
the | back 74 MicroRNAs (miRNAs) |
front 75 95.Which of the following RNAs is not involved in coding for proteins
but plays | back 75 Other noncoding RNAs |
front 76 96.Which type of RNA polymerase is responsible for transcribing most rRNA genes? | back 76 RNA polymerase I |
front 77 What does RNA polymerase II primarily transcribe? | back 77 Protein-coding genes and miRNA genes |
front 78 Which of the following is transcribed by RNA polymerase III? | back 78 tRNA genes and 5S rRNA gene |
front 79 Which RNA polymerase is responsible for transcribing noncoding RNAs
such as | back 79 RNA polymerase II |
front 80 What is the function of RNA polymerase I in eukaryotic cells? | back 80 It transcribes most rRNA genes. |
front 81 Which polymerase transcribes the genes for many small RNAs,
including | back 81 RNA polymerase III |
front 82 RNA polymerase II is primarily involved in transcribing genes that encode | back 82 Protein-coding genes and small noncoding RNAs |
front 83 Which polymerase is involved in the transcription of spliceosome-related | back 83 RNA polymerase II |
front 84 Which type of RNA polymerase is responsible for transcribing tRNA genes? | back 84 RNA polymerase III |
front 85 What is the role of RNA polymerase II in the production of noncoding RNAs? | back 85 It transcribes noncoding RNAs such as those for the spliceosome. |
front 86 Which of the following is a key difference between bacterial and eukaryotic | back 86 Bacterial mRNAs are often polycistronic, encoding multiple proteins. |
front 87 What is a characteristic of eukaryotic mRNAs that is absent in bacterial | back 87 Presence of a poly-A tail at the 3’ end |
front 88 How do bacterial mRNAs differ from eukaryotic mRNAs in terms
of | back 88 Bacterial mRNAs do not have a 5’ cap or a poly-A tail. |
front 89 Which of the following is true regarding the structure of eukaryotic mRNAs? | back 89 Eukaryotic mRNAs have a 5’ cap and a 3’ poly-A tail. |
front 90 What is the significance of a 5’ cap in eukaryotic mRNAs? | back 90 It is required for ribosome binding and initiation of translation. |
front 91 How are bacterial and eukaryotic mRNAs different in terms of introns and | back 91 Eukaryotic mRNAs typically contain introns that are spliced out before translation. |
front 92 Where does transcription and translation occur in bacterial cells? | back 92 Both occur simultaneously in the cytoplasm |
front 93 What is the lifespan of bacterial mRNAs compared to eukaryotic mRNAs? | back 93 Bacterial mRNAs are typically degraded rapidly, with a short half-life. |
front 94 In eukaryotic cells, what processing step is involved in converting pre-mRNA to mature mRNA? | back 94 Splicing out of introns and joining of exons |
front 95 What is a polycistronic mRNA? | back 95 An mRNA that codes for multiple proteins, typically found in bacteria |
front 96 What is the first step in the process of translation? | back 96 The ribosome assembles at the start codon of mRNA. |
front 97 Which codon signals the start of translation in most organisms? | back 97 AUG |
front 98 Which of the following molecules brings amino acids to the ribosome during | back 98 Transfer RNA (tRNA) |
front 99 What happens during the elongation phase of translation? | back 99 tRNA anticodons pair with mRNA codons, adding amino acids to the growing peptide chain. |
front 100 Which of the following is the function of the ribosome during translation? | back 100 It catalyzes the formation of peptide bonds between amino acids. |
front 101 What is the role of the E site on the ribosome? | back 101 It binds the tRNA that has released its amino acid and is about to
exit the |
front 102 Which type of bond forms between amino acids during translation? | back 102 Peptide bond |
front 103 What happens when a stop codon is encountered during translation? | back 103 The ribosome disassembles, and the polypeptide chain is released. |
front 104 What is the mechanism of action of tetracycline? | back 104 Prevents the binding of aminoacyl-tRNA to the A site of the ribosome |
front 105 Which antibiotic inhibits bacterial protein synthesis by preventing the transition from initiation to elongation? | back 105 Streptomycin |
front 106 How does chloramphenicol inhibit bacterial protein synthesis? | back 106 Blocks peptidyl transferase reaction on ribosomes |
front 107 Which antibiotic binds to the exit channel of the ribosome and inhibits elongation of the peptide chain? | back 107 Erythromycin |
front 108 What is the target of rifamycin in bacterial cells? | back 108 Inhibition of RNA polymerase |
front 109 What is the function of the trp operon in Escherichia coli? | back 109 To regulate the production of tryptophan |
front 110 How is the trp operon regulated when tryptophan levels are high? | back 110 The trp repressor binds to the operator, preventing transcription. |
front 111 What is the role of tryptophan in the regulation of the trp operon? | back 111 It acts as a co-repressor, enabling the trp repressor to bind the operator. |
front 112 Where does the trp repressor bind when tryptophan levels are high? | back 112 The operator region |
front 113 What happens to the trp operon when tryptophan is scarce in the | back 113 The trp operon is fully transcribed, leading to the synthesis of enzymes for tryptophan production. |
front 114 What role does RNA polymerase play in the trp operon when tryptophan is | back 114 It binds to the promoter and initiates transcription of the trp operon. |
front 115 What is the role of attenuation in the regulation of the trp operon? | back 115 It allows fine-tuning of transcription based on tryptophan levels. |
front 116 How does the leader peptide sequence affect the trp operon in E. coli? | back 116 It forms secondary structures that can terminate transcription prematurely when tryptophan is abundant. |
front 117 137. What happens when the trp repressor is inactivated? | back 117 RNA polymerase binds to the promoter and transcription of the trp operon begins. |
front 118 What does the trp operon code for? | back 118 Enzymes involved in tryptophan biosynthesis |
front 119 What is the primary function of the lac operon in Escherichia coli? | back 119 To regulate the metabolism of lactose |
front 120 Which gene in the lac operon codes for beta-galactosidase, the enzyme that breaks down lactose? | back 120 lacZ |
front 121 What is the role of the lacI gene in the lac operon? | back 121 It encodes the repressor protein that binds to the operator. |
front 122 What happens when lactose is present in the environment of Escherichia | back 122 Lactose binds to the lac repressor, causing it to release from the operator and allowing transcription of the lac operon. |
front 123 In the absence of lactose, what happens to the lac operon? | back 123 The repressor binds to the operator, blocking transcription |
front 124 Which molecule acts as an inducer for the lac operon? | back 124 Lactose |
front 125 What happens to the lac operon when both glucose and lactose are
present | back 125 Transcription of the lac operon is repressed by the presence of glucose through catabolite repression. |
front 126 What is the role of the CAP-cAMP complex in the regulation of the lac | back 126 It enhances the binding of RNA polymerase to the promoter, increasing transcription in the absence of glucose. |
front 127 What is the effect of glucose on the lac operon? | back 127 It decreases the concentration of cAMP, preventing activation of the lac operon. |
front 128 Which of the following conditions would result in maximum expression of the lac operon? | back 128 No glucose and high lactose |
front 129 How do the mechanisms of repression differ between the trp operon and the lac operon in Escherichia coli with respect to their interaction with their repressor proteins and environmental signals? | back 129 The trp repressor requires a co-repressor (tryptophan) to bind to the operator, while the lac repressor binds to the operator in the absence of lactose and is inactivated when lactose is present. |
front 130 The DNA sequence shown below includes the beginning of a sequence that codes for a protein. What would be the result of a mutation that changed the underlined bold C to an A? 5'-ATCATGAATGTAGCACGCATTCACATAAGGTTT-3' | back 130 M-N-V-A-S-I-H-I-R-F |
front 131 In a DNA double helix, _____ | back 131 the two DNA strands run antiparallel |
front 132 Which of the following sequences can fully base-pair with itself? | back 132 5’-GGATATCC-3’ |
front 133 Which of the following best represents the total number of chromosomes found in each of the somatic cells in your body? | back 133 46 |
front 134 Which of the following histone proteins does not form part of the octameric core? | back 134 H1 |
front 135 What is the most likely explanation for the similarity in the coding
sequences | back 135 Mutations in coding sequences are more likely to be deleterious to the organism than mutations in noncoding sequences. |
front 136 Which of the following does not occur before a eukaryotic mRNA is exported from the nucleus? | back 136 The ribosome binds to the mRNA. |
front 137 Why is the lagging strand synthesized discontinuously at the replication fork? | back 137 DNA polymerase can polymerize nucleotides only in the 5′-to-3′ direction. |
front 138 Which chromatin components are not retained when the classic “beads-on-a-string” structure is generated? | back 138 30-nm fiber |
front 139 Which amino acid would you expect a tRNA with the anticodon 5’-CCA-3’ to carry? | back 139 tryptophan |
front 140 Which protein is responsible for the recruitment of splicing factors to the CTD of RNA Polymerase during transcription initiation in eukaryotes? | back 140 snRNP |
front 141 Which of the following correctly identifies the nucleosomal core histones? | back 141 H2A, H2B, H3 and H4 |
front 142 The DNA from two different species can often be distinguished by a difference in the: | back 142 ratio of A + T to G + C. |
front 143 DNA replication is considered semiconservative because: | back 143 Each daughter DNA molecule consists of one strand from the parent DNA molecule and one new strand. |
front 144 Which of the following sequences can fully base-pair with itself? | back 144 5′-AAGCGCTT-3′ |
front 145 If the genome of a bacterium requires about 20 minutes to replicate, how can the genome of the fruit fly be replicated in only three minutes? | back 145 The fruit fly genome contains more origins of replication than the bacterial genome. |
front 146 Which of the following statements is true? | back 146 rRNA contains the catalytic activity that joins amino acids together. |
front 147 In eukaryotes, but not prokaryotes, ribosomes find the start site of translation by: | back 147 scanning along the mRNA from the 5′ end. |
front 148 The distinct characteristics of different cell types in a multicellular organism are produced mainly by the differential regulation of the: | back 148 transcription of genes transcribed by RNA polymerase II. |
front 149 How are most eukaryotic gene regulatory proteins able to affect transcription when their binding sites are far from the promoter? | back 149 By looping out the intervening DNA between their binding site and the promoter |
front 150 Which amino acid would you expect a tRNA with the anticodon 5′-CUU-3′ to carry? | back 150 lysine (lys) |
front 151 What would be the outcome of the Hershey-Chase experiment if protein (and not DNA) was the carrier of the genetic information? | back 151 The infected bacteria would be positive for S35 radioactive labeling, and not P32. |
front 152 What are the major and minor grooves? | back 152 Spaces between the turns of the DNA double helix |
front 153 The complementary strand for the sequence 5’-GCACTTCG-3’ is: | back 153 3’-CGTGAAGC-5’ |
front 154 What happens to genes present in a part of DNA folded into heterochromatin? | back 154 They will not be expressed. |
front 155 DNA replication origin is a segment of DNA that is most likely to be: | back 155 Rich in A-T base pairthe |
front 156 DNA replication is: | back 156 semi-conservative |
front 157 A replication fork is: | back 157 an open site in the DNA double helix where DNA replication occurs |
front 158 The enzyme that digests the primers made by primase is called: | back 158 ribonuclease |
front 159 In their 1953 paper on the double-helical structure of DNA, Watson and Crick famously wrote: “It has not escaped our notice that the specific pairing we have postulated immediately suggests a possible copying mechanism for the genetic material.” What did they mean? | back 159 Each strand in a DNA double helix contains all the information needed to produce a complementary partner strand. |
front 160 When are chromosomes in their most compact form? | back 160 during mitosis |
front 161 Which statement is true about the association of histone proteins and DNA? | back 161 Histone proteins have a high proportion of positively charged amino acids, which bind tightly to the negatively charged DNA backbone. |
front 162 Where is heterochromatin not commonly located? | back 162 chromosomal regions carrying genes that encode ribosomal proteins |
front 163 Consider the process that a cell uses to replicate its double-stranded DNA before undergoing cell division. Which statement describes the DNA in the resulting daughter cells? | back 163 The double helix in each daughter cell consists of one parent strand and one newly synthesized strand. |
front 164 When is homologous recombination, which can flawlessly repair double-strand DNA breaks, most likely to occur? | back 164 after the cell’s DNA has been replicated |
front 165 DNA mismatch repair typically corrects what percentage of replication errors? | back 165 99% |
front 166 Which of the following best defines a mutation? | back 166 permanent change in a DNA sequence |
front 167 What is true of eukaryotic mRNAs? | back 167 They are translated after they are exported from the nucleus. |
front 168 An RNA chain is synthesized in which direction? | back 168 5′-to-3′ only. |
front 169 In eukaryotes, which parts of a gene are transcribed into RNA? | back 169 introns and exons. |
front 170 How many different aminoacyl-tRNA synthetases do most organisms have? | back 170 one for each amino acid |
front 171 It is thought that, early in the history of life, RNAs not only stored information but acted as catalysts in cells. Which reaction is catalyzed by a ribozyme in present-day cells? | back 171 peptide bond formation during translation |
front 172 Which of the following statements is NOT true about liver cells and kidney cells in the same organism? | back 172 They contain different genes. |
front 173 In eukaryotes, where do transcription regulators bind? | back 173 RNA sequencing |
front 174 In eukaryotes, where do transcription regulators bind? | back 174 upstream, downstream, or within the genes they control. |
front 175 What is an operon? | back 175 a set of genes transcribed as a single mRNA from a single promoter. |
front 176 Which is NOT an example of epigenetic inheritance? | back 176 the inheritance of a single point mutation in a gene. |
front 177 Which does NOT affect gene expression at the post-transcriptional level? | back 177 DNA methylation. |
front 178 The chromatin structure of chromosomal centromeric regions is: | back 178 heterochromatic. |
front 179 The nucleotide sequence of one DNA strand of a DNA double helix
is: | back 179 5ʹ-TGATTGTGGACAAAAATCC-3ʹ |
front 180 In the DNA of certain bacterial cells, 13% of the nucleotides contain adenine. What are the percentages of the other nucleotides? | back 180 A = 13%, T = 13%, C = 37%, G = 37% |
front 181 Which of the following statements are correct? | back 181 many origins of replication, two telomeres, one centromere |
front 182 Which of the following statements are correct? | back 182
|
front 183 You have a segment of DNA that contains the following
sequence: | back 183 The Crick strand is the template strand; RNA pol. moves along the 3' to 5' direction of the template strand. |
front 184 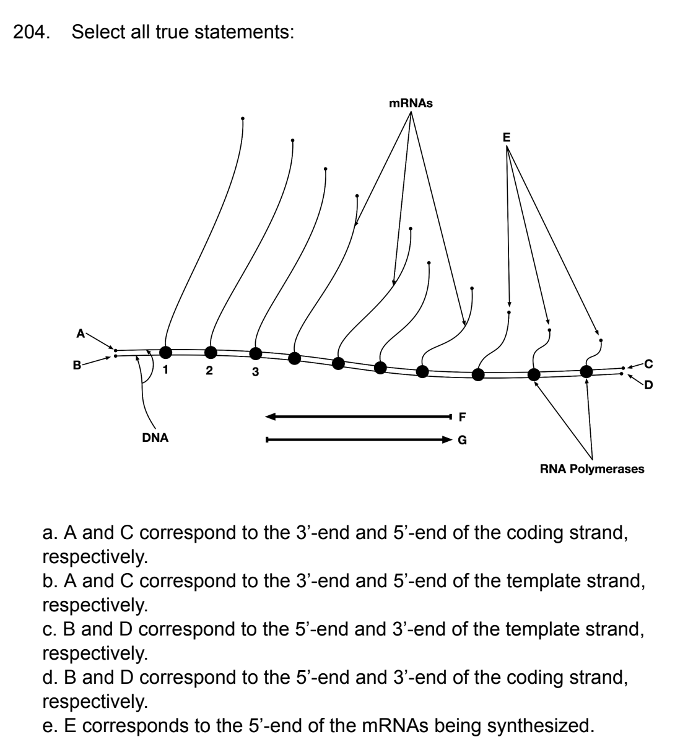 | back 184
|
front 185 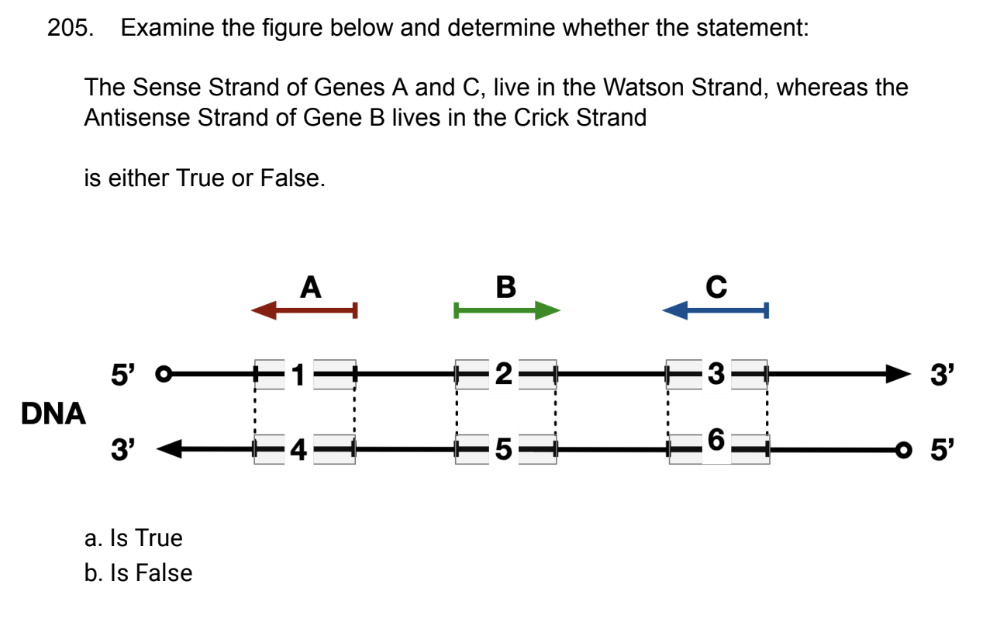 | back 185 false |
front 186 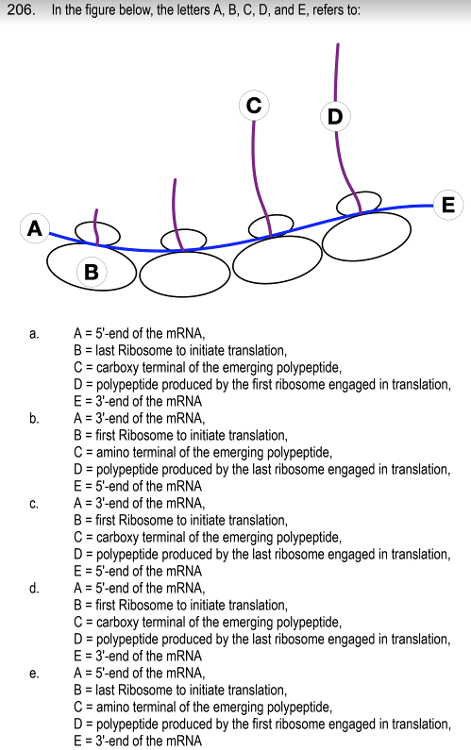 | back 186 A = 5'-end of the mRNA, |
front 187 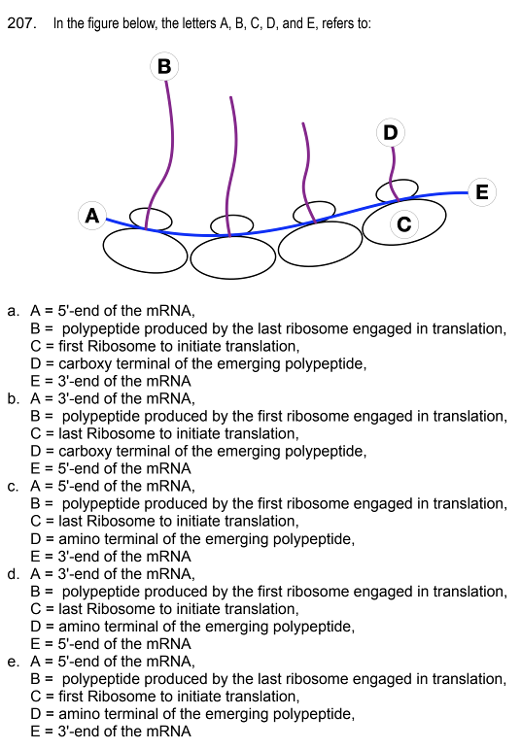 | back 187 A = 3'-end of the mRNA |