

In a DNA sequence, the purine adenine always pairs with the pyrimidine thymine, and the purine guanine always pairs with the pyrimidine cytosine.
What is the difference between purines and pyrimidines?
The complementary partners in a DNA base pair are always made up of one purine and one pyrimidine, never two purines or two pyrimidines. Purines are the nucleotides that have the double-ring structures. Pyrimidines are the nucleotides that have the single-ring structures. An easy way to think of it is that the longer names (pyrimidines) have the smaller rings, and the shorter name (purines) have the bigger rings.


Hint 1.The definition of a complementary sequenceEach of the four nucleotides found in DNA has one complementary partner. Adenine (A) always pairs with thymine (T) and guanine (G) always pairs with cytosine (C) on the complementary DNA strand. The two complementary strands always pair up, each strand running in opposite directions. Below is an example of a DNA sequence and its complement.
Sequence5′−ATGCTACG−3′Complement3′−TACGATGC−5′


Hint 1. Hydrogen bonds and base-pair stabilityIn general, the stability of a base pair is determined by the number of hydrogen bonds between the two bases. The more hydrogen bonds between the bases, the more stable the base pair is.
The base pair adenine-cytosine occurs very rarely in nature. It only happens during a mutation event. When the DNA is replicated, one of the two daughters will contain a guanine-cytosine base pair in the location of the mutation, and the other daughter will contain an adenine-thymine base pair.
If you measure the amounts of the bases in any sample of DNA, you'll find that ...
A. the amount of A + the amount of C = the amount of G + the amount of U.
B. the amount of A + the amount of T = the amount of G + the amount of C.
C. the amount of A + the amount of G = the amount of T + the amount of C.
D. the amount of A = the amount of G; the amount of T = the amount of C.
E. None of the above.
C. the amount of A + the amount of G = the amount of T + the amount of C.
For every A there's a T; for every G there's a C.
Which statement expresses a real difference between DNA and RNA?
A. DNA can make a double helix; RNA cannot.
B. DNA is much longer than RNA.
C. DNA has more kinds of bases than RNA.
D Both (A) and (D).
E. (A), (B), and (C).
B. DNA is much longer than RNA.
RNA is made by copying short segments of DNA.
What holds the strands of the DNA double helix together?
A. Attractions between the ionized phosphates.
B.Attractions between polar sugars.
C. Covalent bonds between A and T, and between C and G.
D. Proteins that bind to DNA bases.
E. Hydrogen bonds between purines and pyrimidines.
E. Hydrogen bonds between purines and pyrimidines.
A and G are purines; C and T are pyrimidines. Hydrogen bonds link them.
If you were to analyze a DNA sample from a eukaryotic cell, you would find that:
A. the number of purines equals the number of pyrimidines.
B. the number of adenines equals the number of uracils.
C. the number of guanines equals the number of cytosines.
D. Both (A) and (C).
E. (A), (B), and (C).
D. Both (A) and (C).
These are consequences of the base-pairing rules, which are fundamental to life.
The building blocks or monomers of nucleic acid molecules are called _____.
A. fatty acids
B. pyrimidines and purines
C. polysaccharides
D. nucleotides
E. DNA and RNA
D. nucleotides
A nucleotide is a nucleic acid monomer consisting of a nitrogen base, a pentose sugar, and a phosphate group. Nucleotides joined together by covalent bonds called phosphodiester linkages form nucleic acid molecules.
The flow of genetic information in a cell goes from _____.
A. protein to RNA to DNA
B. DNA to RNA to protein
C. DNA to lipid to protein
D. DNA to RNA to glucose
E. DNA to ribosomes to RNA
B. DNA to RNA to protein
The information in DNA is transcribed into RNA and then translated into protein.
When nucleotides polymerize to form a nucleic acid _____.
A. covalent bonds form between the bases of two nucleotides
B. a covalent bond forms between the sugar of one nucleotide and the phosphate of a second
C. a hydrogen bond forms between the sugar of one nucleotide and the phosphate of a second
D. hydrogen bonds form between the bases of two nucleotides
B. a covalent bond forms between the sugar of one nucleotide and the phosphate of a second

B. is indicating a single nucleotide.
Which of these is a difference between a DNA and an RNA molecule?
A. DNA contains five-carbon sugars, whereas RNA contains six-carbon sugars.
B. DNA contains nitrogenous bases, whereas RNA contains phosphate groups.
C. DNA is usually double-stranded, whereas RNA is usually single-stranded.
D. DNA contains uracil, whereas RNA contains thymine.
E. DNA is a polymer composed of nucleotides, whereas RNA is a polymer composed of nucleic acids.
C. DNA is usually double-stranded, whereas RNA is usually single-stranded.
With some exceptions, DNA is a double-stranded molecule and RNA is a single-stranded molecule.

A. amino acid
B. nucleic acid
C. nucleotide
D. thiol
E. none of the above
C. nucleotide
Nucleotides are composed of a pentose sugar, a nitrogenous base, and a phosphate group.

A. phosphate group
B. sugar
C. nucleotide
D. nitrogenous base
E. none of the above
A. phosphate group
Phosphate groups contain phosphorus.

C. This is a nitrogenous base.

A. phosphate group, not a uracil
B. double-stranded molecule, not a single-stranded molecule
C. sugar with two, and not three, oxygen atoms
D. thymine nitrogenous base, not a uracil nitrogenous base
E. uracil nitrogenous base, not a thymine nitrogenous base
C. sugar with two, and not three, oxygen atoms
DNA nucleotides are composed of deoxyribose sugars, whereas RNA nucleotides are composed of ribose sugars.
Which of these nitrogenous bases is found in DNA but not in RNA?
A. cytosine
B. guanine
C. thymine
D. adenine
E. uracil
C. thymine
DNA contains thymine; RNA does not.

A. B, C, and D
B. A and B
C. C, D, and E
D. B and C
E. A, B, and C
C. C, D, and E
Pyrimidines are single-ring structures.
In a nucleotide, the nitrogenous base is attached to the sugar's _____ carbon and the phosphate group is attached to the sugar's _____ carbon.
A. 1' ... 3'
B. 2' ... 1'
C. 1' ... 5'
D. 1' ... 2'
E. 2' ... 3'
C. 1' ... 5'
The nitrogenous base is attached to the sugar's 1' carbon and the phosphate group is attached to the sugar's 5' carbon.
Nucleic acids are assembled in the _____ direction.
A. 4' to 5'
B. 5' to 1'
C. 5' to 3'
D. 1' to 5'
E. 2' to 3
C. 5' to 3'
New nucleotides are added to the 3' end of a growing polynucleotide.
In a DNA double helix an adenine of one strand always pairs with a(n) _____ of the complementary strand, and a guanine of one strand always pairs with a(n) _____ of the complementary strand.
A. cytosine ... uracil
B. cytosine ... thymine
C. uracil ... cytosine
D. thymine ... cytosine
E. guanine ... adenine
D. thymine ... cytosine
This is referred to as specific base pairing.

A. moneran
B. phage
C. bacterium
D. protist
E. red blood cell
B. phage
This is a T2 phage, a type of phage that infects E. coli.
Who demonstrated that DNA is the genetic material of the T2 phage?
A. Franklin
B. Hershey and Chase
C. Meselson and Stahl
D. Watson and Crick
E. Darwin and Wallace
B. Hershey and Chase
Hershey and Chase did a series of classic experiments demonstrating that DNA is the genetic material of the T2 phage.
The radioactive isotope 32P labels the T2 phage's _____.
A. tail
B. DNA
C. head
D. protein coat
E. base plate
B. DNA
The T2 phage consists of a protein coat and DNA. It is the DNA that contains P.
Hershey and Chase used _____ to radioactively label the T2 phage's proteins.
A. 14C
B. 32P
C. 92U
D. 35S
E. 222Ra
D. 35S
Hershey and Chase used radioactive sulfur to label the phage's proteins.
After allowing phages grown with bacteria in a medium that contained 32P and 35S, Hershey and Chase used a centrifuge to separate the phage ghosts from the infected cell. They then examined the infected cells and found that they contained _____, which demonstrated that _____ is the phage's genetic material.
A. labeled DNA ... labeled protein
B. labeled protein ... DNA
C. labeled protein .... protein
D. labeled DNA ... DNA
E. labeled DNA .... protein
D. labeled DNA ... DNA
Since the phage DNA entered the infected cell, it makes sense that DNA is the genetic material.
In his transformation experiments, what did Griffith observe?
A. Mice infected with a pathogenic strain of bacteria can spread the infection to other mice.
B. Infecting mice with nonpathogenic strains of bacteria makes them resistant to pathogenic strains.
C. Mixing a heat-killed pathogenic strain of bacteria with a living nonpathogenic strain can convert some of the living cells into the pathogenic form.
D. Mixing a heat-killed nonpathogenic strain of bacteria with a living pathogenic strain makes the pathogenic strain nonpathogenic.
C. Mixing a heat-killed pathogenic strain of bacteria with a living nonpathogenic strain can convert some of the living cells into the pathogenic form.
Cytosine makes up 42% of the nucleotides in a sample of DNA from an organism. Approximately what percentage of the nucleotides in this sample will be thymine?
A. 58%
B. 42%
C. 8%
D. 16%
C. 8%

DNA Structure and Replication Machinery
DNA is composed of two strands that are bound together, resembling a rope ladder with rigid rungs. This DNA ladder is twisted, forming what is called the double helix. The structure of the DNA double helix depends on the complementary pairing of bases between the two strands.
Replication of DNA requires that the two strands of the helix separate, as shown in the image. New daughter molecules are constructed by the sequential addition of nucleotides and the formation of base pairs between the new strand and the parent (template) strand. The replication of the double helix results in two daughter molecules, each composed of one parent strand and one new strand. The enzymes that accomplish the replication of DNA are called DNA polymerases.
NO QUESTION

The DNA double helix is composed of two strands of DNA; each strand is a polymer of DNA nucleotides. Each nucleotide consists of a sugar, a phosphate group, and one of four nitrogenous bases. The structure and orientation of the two strands are important to understanding DNA replication.Drag the labels to their appropriate locations on the diagram below. Pink labels can be used more than once.

Hint 1.Distinguishing the 3' and 5' ends of a DNA strandA strand of DNA consists of a linear polymer of DNA nucleotides. The "backbone" of the DNA strand consists of a repeating pattern of sugar and phosphate groups in which the phosphate of one nucleotide is covalently attached to the sugar of the next nucleotide. This sugar-phosphate-sugar arrangement is called a phosphodiester linkage.
The two ends of a DNA strand are distinct from each other. The 5' end has a phosphate group, which is attached to the 5' carbon of a deoxyribose sugar. At the 3' end, there is a hydroxyl (-OH) group attached to the 3' carbon of a deoxyribose sugar.
Hint 2. What are the components of a DNA strand?A strand of DNA consists of a sequence of covalently linked DNA nucleotides.Drag the terms on the left to the appropriate blanks on the right to complete the sentences.
---
The DNA double helix is constructed from two strands of DNA, each with a sugar-phosphate backbone and nitrogenous bases that form hydrogen bonds, holding the two strands together. Each DNA strand has two unique ends. The 3' end has a hydroxyl (-OH) group on the deoxyribose sugar, whereas the 5' end has a phosphate group. In the double helix, the two strands are antiparallel, that is, they run in opposite directions such that the 3' end of one strand is adjacent to the 5' end of the other strand.
To which end of a primer does DNA polymerase add new nucleotides?
In bacteria, DNA polymerase III is the enzyme that adds new nucleotides to a primer or growing strand of DNA.
A. The newly added nucleotide forms a bond with the phosphate group on the 3' end of the primer.
B. The newly added nucleotide forms a bond with the hydroxyl (-OH) group on the 5' end of the primer.
C. The newly added nucleotide forms a bond with the hydroxyl (-OH) group on the 3' end of the primer.
D. The newly added nucleotide forms a bond with the phosphate group on the 5' end of the primer.
C. The newly added nucleotide forms a bond with the hydroxyl (-OH) group on the 3' end of the primer.

In DNA replication in bacteria, the enzyme DNA polymerase III (abbreviated DNA pol III) adds nucleotides to a template strand of DNA. But DNA pol III cannot start a new strand from scratch. Instead, a primer must pair with the template strand, and DNA pol III then adds nucleotides to the primer, complementary to the template strand. Each of the four images below shows a strand of template DNA (dark blue) with an RNA primer (red) to which DNA pol III will add nucleotides.
In which image will adenine (A) be the next nucleotide to be added to the primer?

In the example above, DNA pol III would add an adenine nucleotide to the 3' end of the primer, where the template strand has thymine as the next available base. You can tell which end is the 3' end by the presence of a hydroxyl (-OH) group.
The structure of DNA polymerase III is such that it can only add new nucleotides to the 3' end of a primer or growing DNA strand. This is because the phosphate group at the 5' end of the new strand and the 3' -OH group on the nucleoside triphosphate will not both fit in the active site of the polymerase.

The replication bubble and antiparallel elongation
DNA replication always begins at an origin of replication. In bacteria, there is a single origin of replication on the circular chromosome, as shown in the image here. Beginning at the origin of replication, the two parental strands (dark blue) separate, forming a replication bubble. At each end of the replication bubble is a replication fork where the parental strands are unwound and new daughter strands (light blue) are synthesized. Movement of the replication forks away from the origin expands the replication bubble until two identical chromosomes are ultimately produced.
NO QUESTION

What will be the directionality of the two new DNA strands?
This image shows a replication bubble in a bacterial chromosome. The region enclosed by the box includes the two parental DNA strands (dark blue) and the two newly synthesized strands (light blue). Each of the images below shows the same four segments of DNA.
Which of the following correctly represents the directionality of the two new DNA strands (light blue) compared to the two parental strands?


In this activity, you will demonstrate your understanding of antiparallel elongation at the replication forks. Keep in mind that the two strands in a double helix are oriented in opposite directions, that is, they are antiparallel.
Drag the arrows onto the diagram below to indicate the direction that DNA polymerase III moves along the parental (template) DNA strands at each of the two replication forks. Arrows can be used once, more than once, or not at all.

DNA polymerase III can only add nucleotides to the 3' end of a new DNA strand. Because the two parental DNA strands of a double helix are antiparallel (go from 3' to 5' in opposite directions), the direction that DNA pol III moves on each strand emerging from a single replication fork must also be opposite.
For example, in the replication fork on the left, the new strand on top is being synthesized from 5' to 3', and therefore DNA pol III moves away from the replication fork. Similarly, the new strand on the bottom of that same replication fork is being synthesized from 5' to 3'. But because the bottom parental strand is running in the opposite direction of the top parental strand, DNA pol III moves toward the replication fork.
In summary, at a single replication fork, one strand is synthesized away from the replication fork, and one strand is synthesized toward the replication fork. When you look at both replication forks, note that a single new strand is built in the same direction on both sides of the replication bubble.

Unwinding the DNA
As DNA replication continues and the replication bubble expands, the parental double helix is unwound and separated into its two component strands. This unwinding and separating of the DNA requires three different types of proteins: helicase, topoisomerase, and single-strand binding proteins.Sort the phrases into the appropriate bins depending on which protein they describe.

At each replication fork, helicase moves along the parental DNA, separating the two strands by breaking the hydrogen bonds between the base pairs. (This makes the two parental DNA strands available to the DNA polymerases for replication.) As soon as the base pairs separate at the replication fork, single-strand binding proteins attach to the separated strands and prevent the parental strands from rejoining.
As helicase separates the two parental strands, the parental DNA ahead of the replication fork becomes more tightly coiled. To relieve strain ahead of the replication fork, topoisomerase breaks a covalent bond in the sugar-phosphate backbone of one of the two parental strands. Breaking this bond allows the DNA to swivel around the corresponding bond in the other strand and relieves the strain caused by the unwinding of the DNA at the helicase.
How does topoisomerase work?
Topoisomerase is an enzyme that relieves the strain (tighter twisting) caused by the unwinding of parental DNA by the helicase at the replication fork.
Which of the following statements correctly describes how topoisomerase functions?
A. Topoisomerase breaks a covalent bond between a deoxyribose sugar and a nitrogenous base in one parental strand.
B. Topoisomerase breaks covalent bonds in the backbones of both parental strands.
C. Topoisomerase breaks hydrogen bonds between the two parental strands.
D. Topoisomerase breaks a covalent bond in the backbone of one parental strand.
D. Topoisomerase breaks a covalent bond in the backbone of one parental strand.
What is the function of single-strand binding proteins?
Which of the following statements correctly describes the role of single-strand binding proteins in DNA replication?
A. Single-strand binding proteins bind to parental DNA immediately after the helicase, preventing the two single strands from joining and re-forming a double helix.
B. Single-strand binding proteins bind to double-stranded DNA ahead of the replication fork, relieving the strain caused by helicase.
C. Single-strand binding proteins bind to the newly synthesized strand of DNA immediately after the DNA polymerase.
D. Single-strand binding proteins bind to double-stranded DNA, causing the two strands to separate into single strands.
A. Single-strand binding proteins bind to parental DNA immediately after the helicase, preventing the two single strands from joining and re-forming a double helix.

The role of hydrogen bonds in DNA structure
In a DNA double helix, the two strands of DNA are held together by hydrogen bonds between complementary base pairs. When two single complementary strands of DNA are near each other, hydrogen bonding between the bases usually causes the two strands to join and form a double helix.
NO QUESTION

Comparing the leading and lagging strands
As the two parental (template) DNA strands separate at a replication fork, each of the strands is separately copied by a DNA polymerase III (orange), producing two new daughter strands (light blue), each complementary to its respective parental strand. Because the two parental strands are antiparallel, the two new strands (the leading and lagging strands) cannot be synthesized in the same way.
NO QUESTION

In which direction do the leading and lagging strands elongate?
At a replication fork, the two parental DNA strands separate, and each strand is copied by a DNA polymerase III, synthesizing a new, complementary strand. The diagram below shows a replication fork with the two parental DNA strands labeled at their 3' and 5' ends. The newly synthesized DNA strands are not shown, but the polymerase on each parental strand is shown (labeled 1 and 2).
NO QUESTION
Remembering that the new (daughter) DNA strands will run antiparallel to the parental strands, which of the following statements correctly describes the direction in which the two strands will elongate?
A. Polymerase 1 elongates its strand toward the replication fork, but polymerase 2 elongates its strand away from the replication fork.
B. Both strands elongate toward the replication fork.
C. Both strands elongate away from the replication fork.
D. Polymerase 1 elongates its strand away from the replication fork, but polymerase 2 elongates its strand toward the replication fork.
A. Polymerase 1 elongates its strand toward the replication fork, but polymerase 2 elongates its strand away from the replication fork.

The role of RNA primers in DNA replication
DNA polymerase III cannot initiate the synthesis of a new daughter strand; it can only add nucleotides to the 3' end of an existing strand that is paired with a parental (template, dark blue) strand. The enzyme primase creates a short RNA primer (red) that is complementary to the parental strand. This primer serves as a starting point for DNA pol III, which adds nucleotides to the 3' end of the primer. Think about the number of primers needed for a strand that is synthesized continuously versus a strand that is made in many smaller fragments.
NO QUESTION

Synthesis of the first primer on the lagging strand
As a replication bubble forms at an origin of replication, the primers for the leading strands are produced first. In the image below, you can see that the first parts of the leading strands have already been synthesized by the time the first lagging strand primers are produced.
The reason for this lag is that about 1500 nucleotides (the average length of a bacterial lagging strand segment) of parental DNA must be exposed at the replication fork before lagging strand synthesis can begin. This makes room for the first lagging strand segment between the first primer and the origin of replication. DNA polymerase III produces the first lagging strand segments (not shown) by adding nucleotides to the 3' ends of the lagging strand primers.
NO QUESTION
Which primer on the lagging strand was produced last?
As the replication bubble expands, a new primer must be added to initiate each successive segment of the lagging strand.
On either side of the replication bubble, the most recently added RNA primer on the lagging strand will be located:
A. closest to the origin of replication.
B. closest to the replication fork.
B. closest to the replication fork.

Drag each phrase to the appropriate bin depending on whether it describes the synthesis of the leading strand, the synthesis of the lagging strand, or the synthesis of both strands.

Because DNA polymerase III can only add nucleotides to the 3' end of a new DNA strand and because the two parental DNA strands are antiparallel, synthesis of the leading strand differs from synthesis of the lagging strand.
- The leading strand is made continuously from a single RNA primer located at the origin of replication. DNA pol III adds nucleotides to the 3' end of the leading strand so that it elongates toward the replication fork.
- In contrast, the lagging strand is made in segments, each with its own RNA primer. DNA pol III adds nucleotides to the 3' end of the lagging strand so that it elongates away from the replication fork.
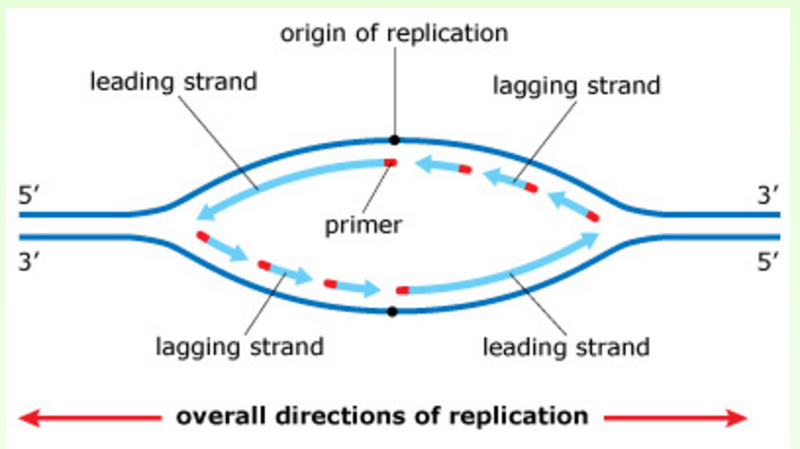
In this image, you can see that on one side of the origin of replication, a new strand is synthesized as the leading strand, and on the other side of the origin of replication, that same new strand is synthesized as the lagging strand. The leading and lagging strands built on the same template strand will eventually be joined, forming a continuous daughter strand.
NO QUESTION

RNA primers on the leading and lagging strands
The diagram below shows a replication bubble with synthesis of the leading and lagging strands on both sides of the bubble. The parental DNA is shown in dark blue, the newly synthesized DNA is light blue, and the RNA primers associated with each strand are red. The origin of replication is indicated by the black dots on the parental strands.
Rank the primers in the order they were produced. If two primers were produced at the same time, overlap them.

As soon as the replication bubble opens and the replication machinery is assembled at the two replication forks, the two primers for the leading strands (primers a and h) are produced. The production of the first primers on the lagging strands (those closest to the origin of replication, b and g) is delayed slightly because the replication forks must open up further to expose the template DNA for the lagging strands.
After completion of the first segments of the lagging strands, additional template DNA must be exposed before the second primers (c and f) can be produced. And after completion of the second segments, additional template DNA must be exposed before the third primers (d and e) can be produced. In summary, because of the way the replication bubble expands, the lagging strand primers near the origin of replication were produced before the primers near the replication forks.

Synthesis of the lagging strand
In contrast to the leading strand, the lagging strand is synthesized as a series of segments called Okazaki fragments. The diagram below illustrates a lagging strand with the replication fork off-screen to the right. Fragment A is the most recently synthesized Okazaki fragment. Fragment B will be synthesized next in the space between primers A and B.
NO QUESTION

Synthesis of the lagging strand is accomplished through the repetition of the following steps.
- Step 1: A new fragment begins with DNA polymerase III binding to the 3' end of the most recently produced RNA primer, primer B in this case, which is closest to the replication fork. DNA pol III then adds DNA nucleotides in the 5' to 3' direction until it encounters the previous RNA primer, primer A.
- Step 2: DNA pol III falls off and is replaced by DNA pol I. Starting at the 5' end of primer A, DNA pol I removes each RNA nucleotide and replaces it with the corresponding DNA nucleotide. (DNA pol I adds the nucleotides to the 3' end of fragment B.) When it encounters the 5' end of fragment A, DNA pol I falls off, leaving a gap in the sugar-phosphate backbone between fragments A and B.
- Step 3: DNA ligase closes the gap between fragments A and B.
These steps will be repeated as the replication fork opens up. Try to visualize primer C being produced to the right (closest to the replication fork). Fragment C would be synthesized and joined to fragment B following the steps described here.
NO QUESTION
In bacteria, the three primary enzymes involved in lagging strand synthesis are DNA polymerase III, DNA polymerase I, and DNA ligase.
Select the two phrases that correctly describe the functions of DNA polymerase I.
A. binds to the 3' end of an RNA primer
B. replaces the RNA nucleotides of primers with DNA nucleotides
C. leaves a gap in the sugar-phosphate backbone after replacing the last RNA nucleotide of a primer
D. joins the sugar-phosphate backbone between a new DNA fragment and the previous fragment
B & C

Drag the labels to their appropriate locations in the flowchart below, indicating the sequence of events in the production of fragment B. (Note that pol I stands for DNA polymerase I, and pol III stands for DNA polymerase III.)

Replication in prokaryotes differs from replication in eukaryotes for which of the following reasons?
A. The rate of elongation during DNA replication is slower in prokaryotes than in eukaryotes.
B. Prokaryotic chromosomes have histones, whereas eukaryotic chromosomes do not.
C. Prokaryotes produce Okazaki fragments during DNA replication, but eukaryotes do not.
D. Prokaryotic chromosomes have a single origin of replication, whereas eukaryotic chromosomes have many.
D. Prokaryotic chromosomes have a single origin of replication, whereas eukaryotic chromosomes have many.
In E. coli, there is a mutation in a gene called dnaB that alters the helicase that normally acts at the origin. Which of the following would you expect as a result of this mutation?
A. Replication will require a DNA template from another source.
B. Additional proofreading will occur.
C. Replication will occur via RNA polymerase alone.
D. No replication fork will be formed.
D. No replication fork will be formed.
How does the enzyme telomerase meet the challenge of replicating the ends of linear chromosomes?
A. It adds numerous GC pairs, which resist hydrolysis and maintain chromosome integrity.
B. It causes specific double-strand DNA breaks that result in blunt ends on both strands.
C. It adds a single 5' cap structure that resists degradation by nucleases.
D. It catalyzes the lengthening of telomeres, compensating for the shortening that could occur during replication without telomerase activity.
D. It catalyzes the lengthening of telomeres, compensating for the shortening that could occur during replication without telomerase activity.
What is the role of DNA ligase in the elongation of the lagging strand during DNA replication?
A. It synthesizes RNA nucleotides to make a primer.
B. It stabilizes the unwound parental DNA.
C. It unwinds the parental double helix.
D. It joins Okazaki fragments together.
D. It joins Okazaki fragments together.
Which of the following help(s) to hold the DNA strands apart while they are being replicated?
A. primase
B. ligase
C. single-strand DNA binding proteins
D. DNA polymerase
C. single-strand DNA binding proteins
Which of the following would you expect of a eukaryote lacking telomerase?
A. an inability to repair thymine dimers
B. an inability to produce Okazaki fragments
C. a high probability of somatic cells becoming cancerous
D. a reduction in chromosome length in gametes
D. a reduction in chromosome length in gametes

In the late 1950s, Meselson and Stahl grew bacteria in a medium containing "heavy" nitrogen (15N) and then transferred them to a medium containing 14N. Which of the results in the figure above would be expected after one round of DNA replication in the presence of 14N?
D.
If a cell were unable to produce histone proteins, which of the following would be a likely effect?
A. Amplification of other genes would compensate for the lack of histones.
B. Spindle fibers would not form during prophase.
C. The cell's DNA couldn't be packed into its nucleus.
D. There would be an increase in the amount of "satellite" DNA produced during centrifugation.
C. The cell's DNA couldn't be packed into its nucleus.
Why do histones bind tightly to DNA?
A. Histones are negatively charged, and DNA is positively charged.
B. Histones are positively charged, and DNA is negatively charged.
C. Histones are covalently linked to the DNA.
D. Both histones and DNA are strongly hydrophobic.
B. Histones are positively charged, and DNA is negatively charged.
Which of the following statements describes chromatin?
A. Euchromatin is not transcribed, whereas heterochromatin is transcribed.
B. Heterochromatin is highly condensed, whereas euchromatin is less compact.
C. Heterochromatin is composed of DNA, whereas euchromatin is made of DNA and RNA.
D> Both heterochromatin and euchromatin are found in the cytoplasm.
B. Heterochromatin is highly condensed, whereas euchromatin is less compact.